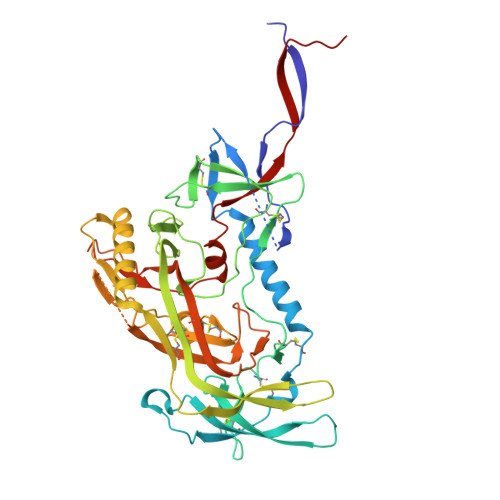
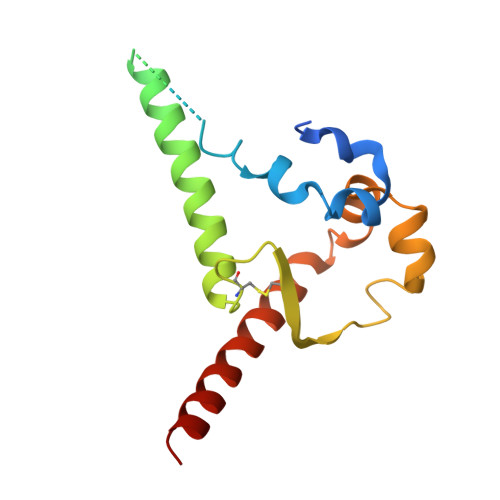
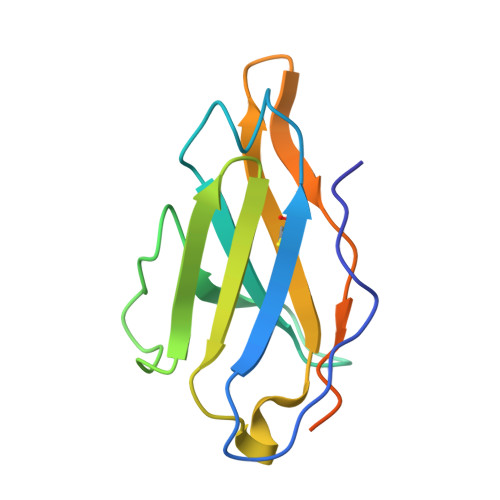
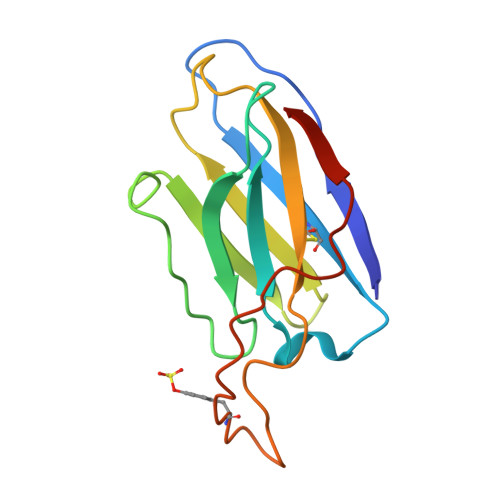
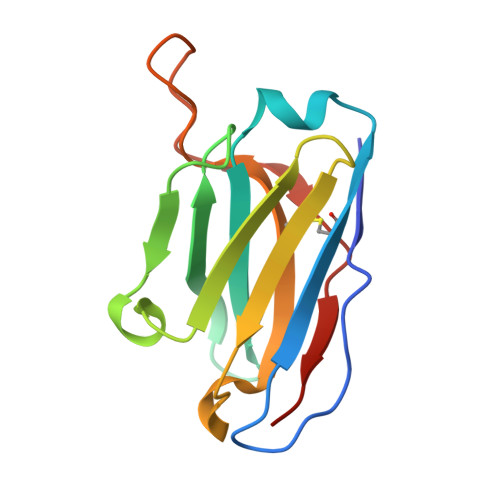
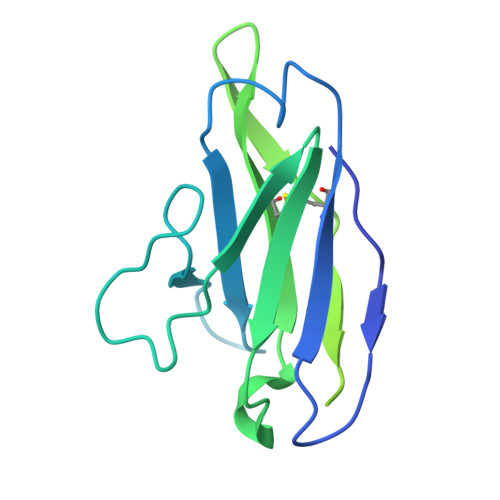
HIV broadly neutralizing antibody precursors to the Apex epitope induced in nonhuman primates.
Ma, K.M., Sutton, H.J., Pratap, P.P., Steichen, J.M., Carnathan, D., Quinn, J., Kalyuzhniy, O., Liguori, A., Agrawal, S., Baboo, S., Madden, P., Cottrell, C.A., Willis, J.R., Lee, J.H., Landais, E., Hu, X., Ramezani-Rad, P., Ozorowski, G., Lewis, V.R., Diedrich, J.K., Zhou, X., Altheide, T.K., Phelps, N., Georgeson, E., Alavi, N.B., Lu, D., Eskandarzadeh, S., Kubitz, M., Adachi, Y., Mullen, T.M., Silva, M., Melo, M.B., Himansu, S., Irvine, D.J., Burton, D.R., Yates 3rd, J.R., Paulson, J.C., Sok, D., Wilson, I.A., Silvestri, G., Ward, A.B., Crotty, S., Schief, W.R.(2025) Sci Immunol 10: eadt6660-eadt6660
- PubMed: 40845127
- DOI: https://doi.org/10.1126/sciimmunol.adt6660
- Primary Citation of Related Structures:
9B8B, 9B8C, 9MPB, 9MPC, 9MPX, 9MQG - PubMed Abstract:
An effective prophylactic HIV vaccine will likely need to induce broadly neutralizing antibodies (bnAbs). bnAbs to the Apex region of the HIV envelope glycoprotein (Env) are promising targets for vaccination because of their relatively low somatic hypermutation compared with other bnAbs. Most Apex bnAbs engage Env using an exceptionally long heavy-chain complementarity-determining region 3 (HCDR3) containing specific binding motifs, which reduces bnAb precursor frequency and makes priming of rare bnAb precursors a likely limiting step in the path to Apex bnAb induction. We found that adjuvanted protein or mRNA lipid nanoparticle (LNP) immunization of rhesus macaques with ApexGT6, an Env trimer engineered to bind Apex bnAb precursors, consistently induced Apex bnAb-related precursors with long HCDR3s bearing bnAb-like sequence motifs. Cryo-electron microscopy revealed that elicited Apex bnAb-related HCDR3s had structures combining elements of several prototype Apex bnAbs. These results achieve a critical HIV vaccine development milestone in outbred primates.
- Consortium for HIV/AIDS Vaccine Development (CHAVD), Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: