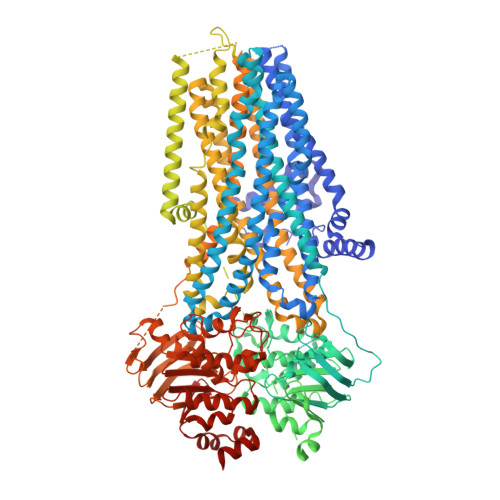
Structural basis for CFTR inhibition by CFTR inh -172.
Young, P.G., Levring, J., Fiedorczuk, K., Blanchard, S.C., Chen, J.(2024) Proc Natl Acad Sci U S A 121: e2316675121-e2316675121
- PubMed: 38422021
- DOI: https://doi.org/10.1073/pnas.2316675121
- Primary Citation of Related Structures:
8UBR - PubMed Abstract:
The cystic fibrosis transmembrane conductance regulator (CFTR) is an anion channel that regulates electrolyte and fluid balance in epithelial tissues. While activation of CFTR is vital to treating cystic fibrosis, selective inhibition of CFTR is a potential therapeutic strategy for secretory diarrhea and autosomal dominant polycystic kidney disease. Although several CFTR inhibitors have been developed by high-throughput screening, their modes of action remain elusive. In this study, we determined the structure of CFTR in complex with the inhibitor CFTR inh -172 to an overall resolution of 2.7 Å by cryogenic electron microscopy. We observe that CFTR inh -172 binds inside the pore near transmembrane helix 8, a critical structural element that links adenosine triphosphate hydrolysis with channel gating. Binding of CFTR inh -172 stabilizes a conformation in which the chloride selectivity filter is collapsed, and the pore is blocked from the extracellular side of the membrane. Single-molecule fluorescence resonance energy transfer experiments indicate that CFTR inh -172 inhibits channel gating without compromising nucleotide-binding domain dimerization. Together, these data reconcile previous biophysical observations and provide a molecular basis for the activity of this widely used CFTR inhibitor.
- Laboratory of Membrane Biology and Biophysics, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: