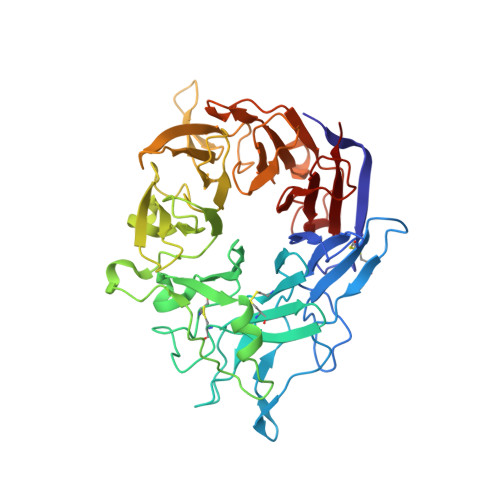
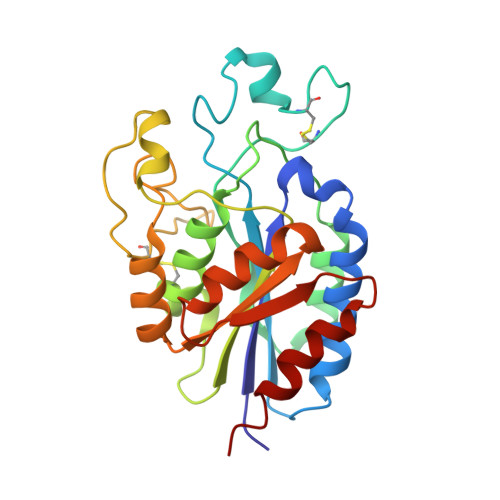
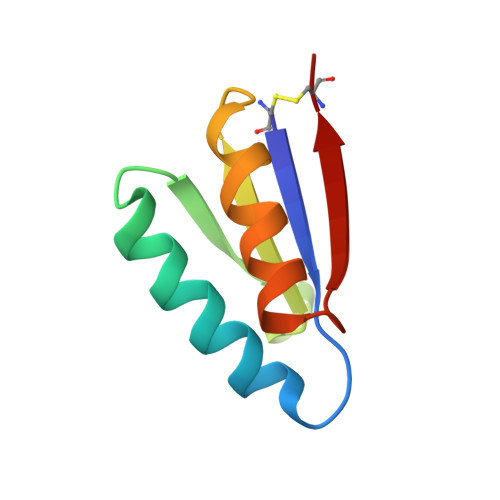
De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Roy, A., Shi, L., Chang, A., Dong, X., Fernandez, A., Kraft, J.C., Li, J., Le, V.Q., Winegar, R.V., Cherf, G.M., Slocum, D., Poulson, P.D., Casper, G.E., Vallecillo-Zuniga, M.L., Valdoz, J.C., Miranda, M.C., Bai, H., Kipnis, Y., Olshefsky, A., Priya, T., Carter, L., Ravichandran, R., Chow, C.M., Johnson, M.R., Cheng, S., Smith, M., Overed-Sayer, C., Finch, D.K., Lowe, D., Bera, A.K., Matute-Bello, G., Birkland, T.P., DiMaio, F., Raghu, G., Cochran, J.R., Stewart, L.J., Campbell, M.G., Van Ry, P.M., Springer, T., Baker, D.(2023) Nat Commun 14: 5660-5660
- PubMed: 37704610
- DOI: https://doi.org/10.1038/s41467-023-41272-z
- Primary Citation of Related Structures:
7LMV, 7LMX, 8TCF, 8TCG - PubMed Abstract:
The RGD (Arg-Gly-Asp)-binding integrins αvβ6 and αvβ8 are clinically validated cancer and fibrosis targets of considerable therapeutic importance. Compounds that can discriminate between homologous αvβ6 and αvβ8 and other RGD integrins, stabilize specific conformational states, and have high thermal stability could have considerable therapeutic utility. Existing small molecule and antibody inhibitors do not have all these properties, and hence new approaches are needed. Here we describe a generalized method for computationally designing RGD-containing miniproteins selective for a single RGD integrin heterodimer and conformational state. We design hyperstable, selective αvβ6 and αvβ8 inhibitors that bind with picomolar affinity. CryoEM structures of the designed inhibitor-integrin complexes are very close to the computational design models, and show that the inhibitors stabilize specific conformational states of the αvβ6 and the αvβ8 integrins. In a lung fibrosis mouse model, the αvβ6 inhibitor potently reduced fibrotic burden and improved overall lung mechanics, demonstrating the therapeutic potential of de novo designed integrin binding proteins with high selectivity.
- Department of Biochemistry and Institute for Protein Design, University of Washington, Seattle, WA, 98195, USA.
Organizational Affiliation: