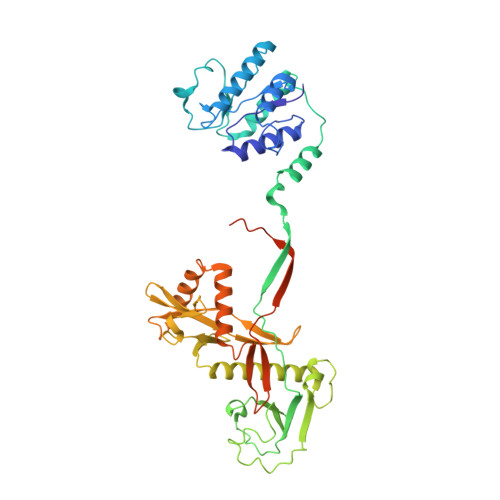
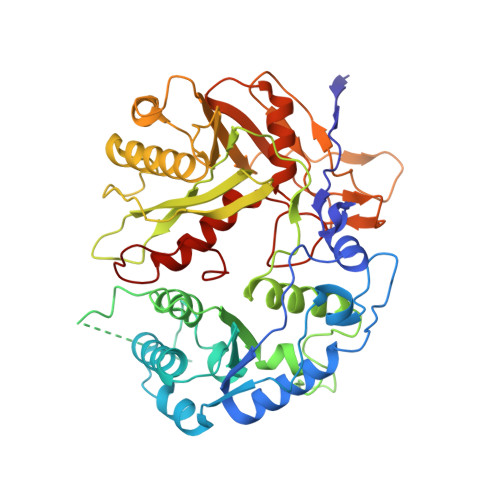
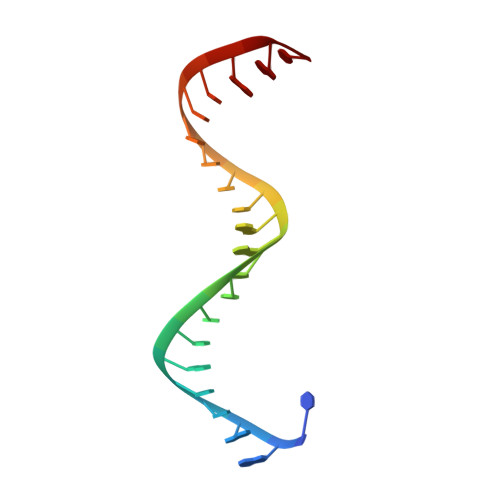
Oligomerization-mediated activation of a short prokaryotic Argonaute.
Shen, Z., Yang, X.Y., Xia, S., Huang, W., Taylor, D.J., Nakanishi, K., Fu, T.M.(2023) Nature 621: 154-161
- PubMed: 37494956
- DOI: https://doi.org/10.1038/s41586-023-06456-z
- Primary Citation of Related Structures:
8FEX, 8FFI, 8SP0, 8SP3, 8SPO, 8SQU - PubMed Abstract:
Although eukaryotic and long prokaryotic Argonaute proteins (pAgos) cleave nucleic acids, some short pAgos lack nuclease activity and hydrolyse NAD(P) + to induce bacterial cell death 1 . Here we present a hierarchical activation pathway for SPARTA, a short pAgo consisting of an Argonaute (Ago) protein and TIR-APAZ, an associated protein 2 . SPARTA progresses through distinct oligomeric forms, including a monomeric apo state, a monomeric RNA-DNA-bound state, two dimeric RNA-DNA-bound states and a tetrameric RNA-DNA-bound active state. These snapshots together identify oligomerization as a mechanistic principle of SPARTA activation. The RNA-DNA-binding channel of apo inactive SPARTA is occupied by an auto-inhibitory motif in TIR-APAZ. After the binding of RNA-DNA, SPARTA transitions from a monomer to a symmetric dimer and then an asymmetric dimer, in which two TIR domains interact through charge and shape complementarity. Next, two dimers assemble into a tetramer with a central TIR cluster responsible for hydrolysing NAD(P) + . In addition, we observe unique features of interactions between SPARTA and RNA-DNA, including competition between the DNA 3' end and the auto-inhibitory motif, interactions between the RNA G2 nucleotide and Ago, and splaying of the RNA-DNA duplex by two loops exclusive to short pAgos. Together, our findings provide a mechanistic basis for the activation of short pAgos, a large section of the Ago superfamily.
- Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH, USA.
Organizational Affiliation: