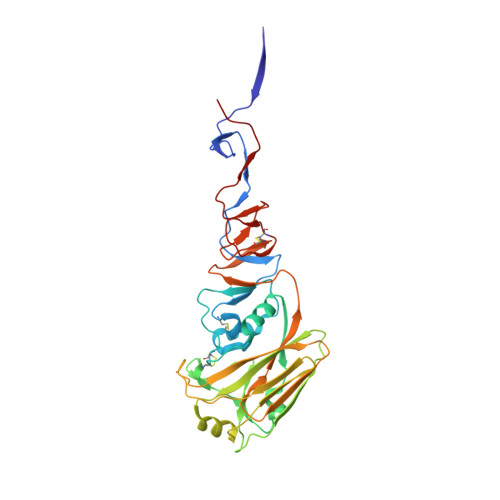
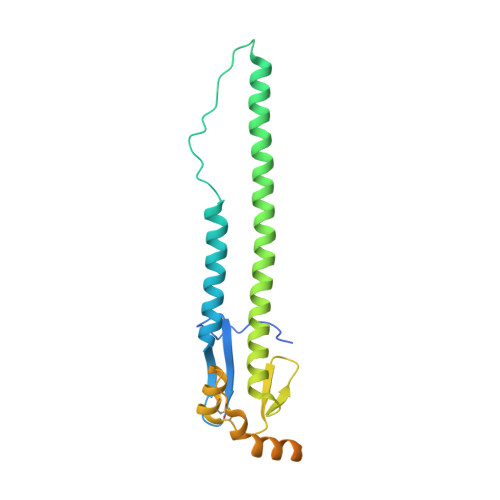
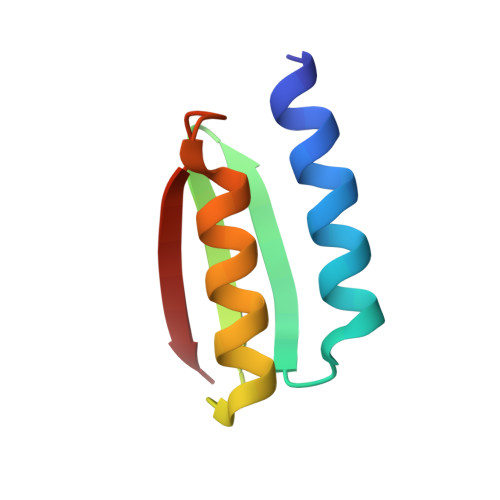
De novo design of protein structure and function with RFdiffusion.
Watson, J.L., Juergens, D., Bennett, N.R., Trippe, B.L., Yim, J., Eisenach, H.E., Ahern, W., Borst, A.J., Ragotte, R.J., Milles, L.F., Wicky, B.I.M., Hanikel, N., Pellock, S.J., Courbet, A., Sheffler, W., Wang, J., Venkatesh, P., Sappington, I., Torres, S.V., Lauko, A., De Bortoli, V., Mathieu, E., Ovchinnikov, S., Barzilay, R., Jaakkola, T.S., DiMaio, F., Baek, M., Baker, D.(2023) Nature 620: 1089-1100
- PubMed: 37433327
- DOI: https://doi.org/10.1038/s41586-023-06415-8
- Primary Citation of Related Structures:
8SK7 - PubMed Abstract:
There has been considerable recent progress in designing new proteins using deep-learning methods 1-9 . Despite this progress, a general deep-learning framework for protein design that enables solution of a wide range of design challenges, including de novo binder design and design of higher-order symmetric architectures, has yet to be described. Diffusion models 10,11 have had considerable success in image and language generative modelling but limited success when applied to protein modelling, probably due to the complexity of protein backbone geometry and sequence-structure relationships. Here we show that by fine-tuning the RoseTTAFold structure prediction network on protein structure denoising tasks, we obtain a generative model of protein backbones that achieves outstanding performance on unconditional and topology-constrained protein monomer design, protein binder design, symmetric oligomer design, enzyme active site scaffolding and symmetric motif scaffolding for therapeutic and metal-binding protein design. We demonstrate the power and generality of the method, called RoseTTAFold diffusion (RFdiffusion), by experimentally characterizing the structures and functions of hundreds of designed symmetric assemblies, metal-binding proteins and protein binders. The accuracy of RFdiffusion is confirmed by the cryogenic electron microscopy structure of a designed binder in complex with influenza haemagglutinin that is nearly identical to the design model. In a manner analogous to networks that produce images from user-specified inputs, RFdiffusion enables the design of diverse functional proteins from simple molecular specifications.
- Department of Biochemistry, University of Washington, Seattle, WA, USA.
Organizational Affiliation: