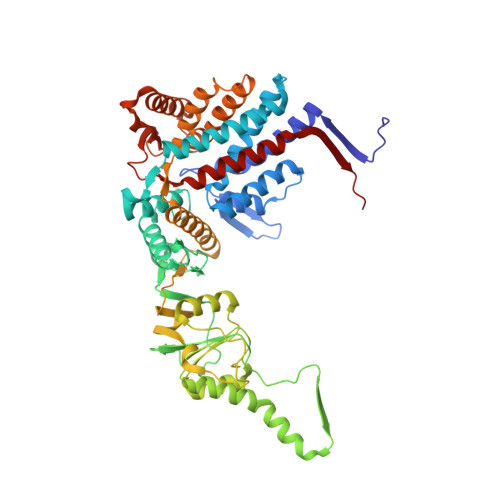
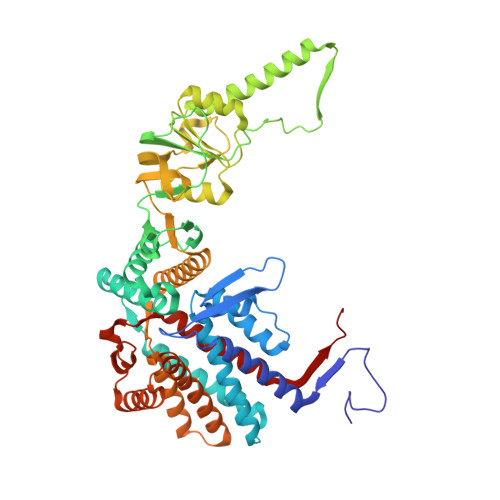
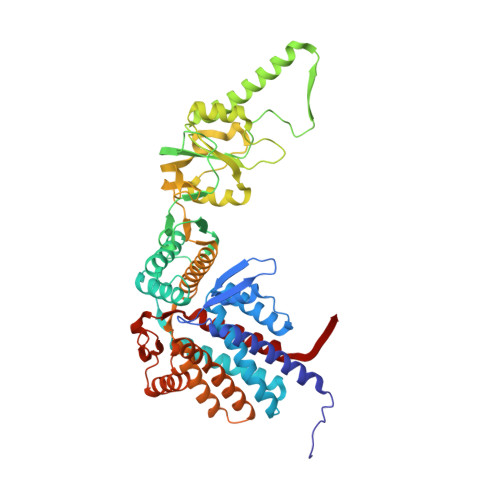
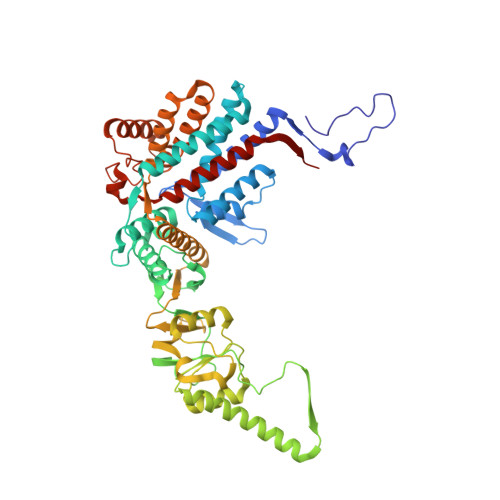
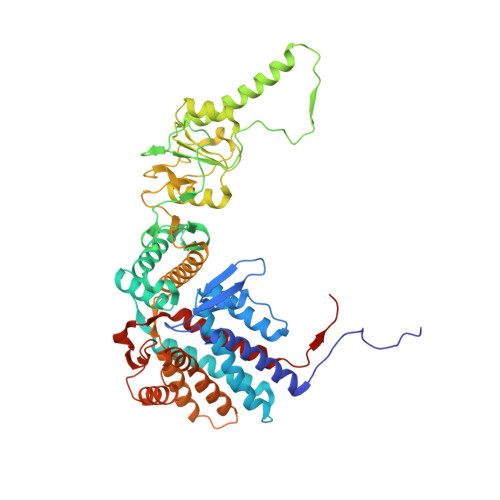
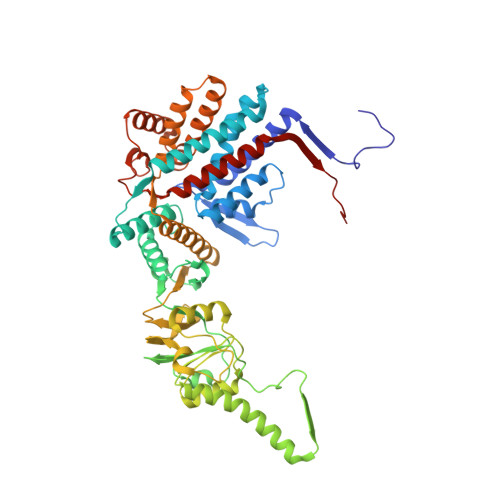
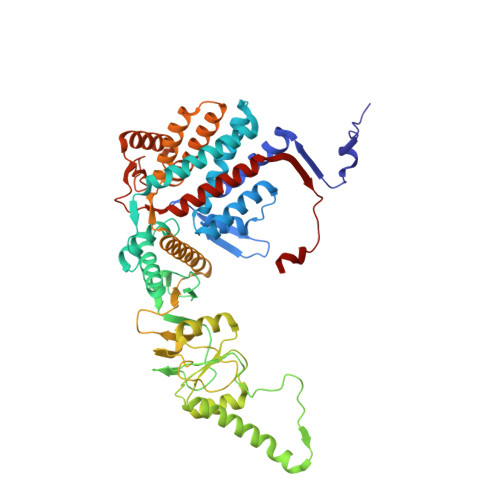
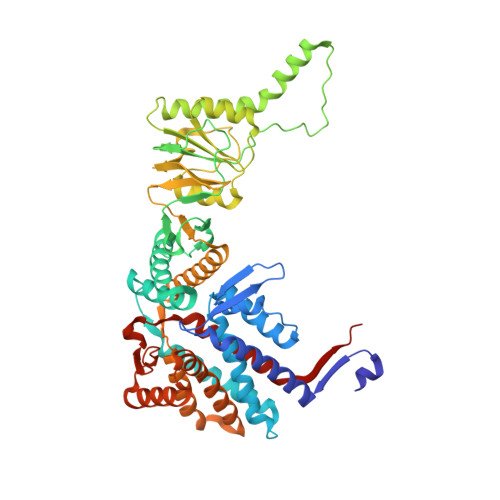
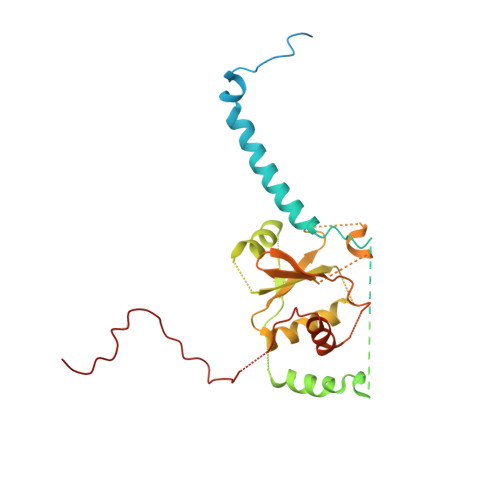
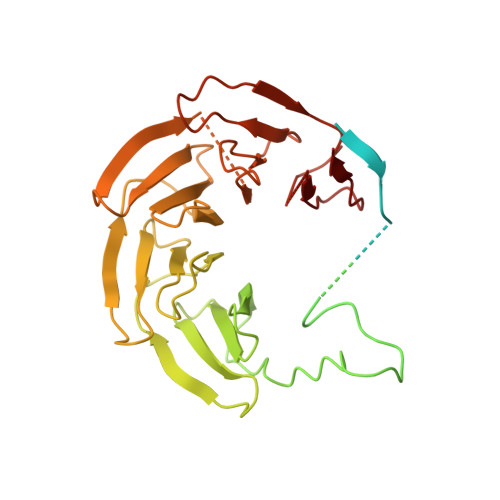
Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Wang, S., Sass, M.I., Kwon, Y., Ludlam, W.G., Smith, T.M., Carter, E.J., Gladden, N.E., Riggi, M., Iwasa, J.H., Willardson, B.M., Shen, P.S.(2023) Mol Cell 83: 3852-3868.e6
- PubMed: 37852256
- DOI: https://doi.org/10.1016/j.molcel.2023.09.032
- Primary Citation of Related Structures:
8SFE, 8SFF, 8SG8, 8SG9, 8SGC, 8SGL, 8SGQ, 8SH9, 8SHA, 8SHD, 8SHE, 8SHF, 8SHG, 8SHL, 8SHN, 8SHO, 8SHP, 8SHQ, 8SHT - PubMed Abstract:
The Chaperonin Containing Tailless polypeptide 1 (CCT) complex is an essential protein folding machine with a diverse clientele of substrates, including many proteins with β-propeller domains. Here, we determine the structures of human CCT in complex with its accessory co-chaperone, phosducin-like protein 1 (PhLP1), in the process of folding Gβ 5 , a component of Regulator of G protein Signaling (RGS) complexes. Cryoelectron microscopy (cryo-EM) and image processing reveal an ensemble of distinct snapshots that represent the folding trajectory of Gβ 5 from an unfolded molten globule to a fully folded β-propeller. These structures reveal the mechanism by which CCT directs Gβ 5 folding through initiating specific intermolecular contacts that facilitate the sequential folding of individual β sheets until the propeller closes into its native structure. This work directly visualizes chaperone-mediated protein folding and establishes that CCT orchestrates folding by stabilizing intermediates through interactions with surface residues that permit the hydrophobic core to coalesce into its folded state.
- Department of Biochemistry, School of Medicine, University of Utah, 15 N. Medical Drive East, Salt Lake City, UT 84112, USA.
Organizational Affiliation: