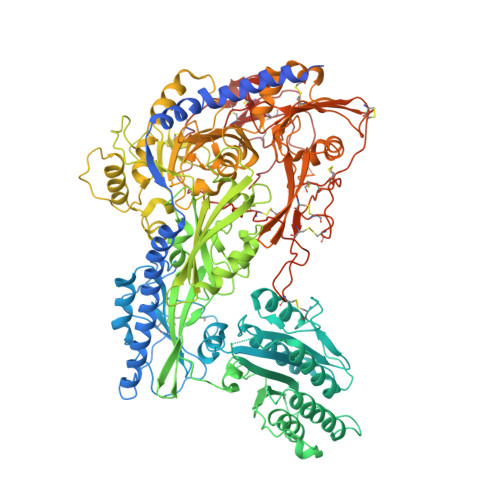
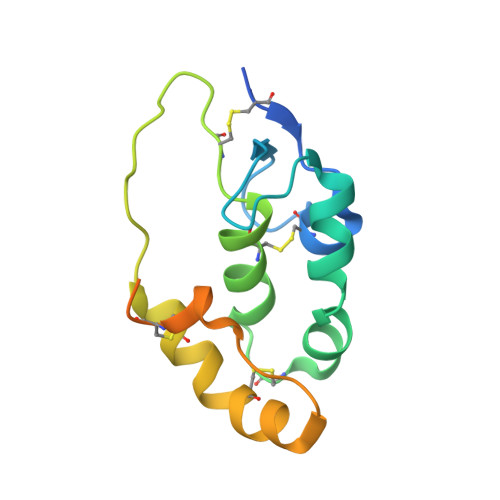
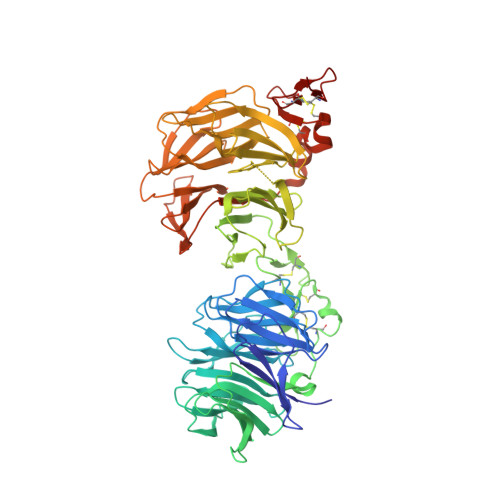
Cachd1 interacts with Wnt receptors and regulates neuronal asymmetry in the zebrafish brain.
Powell, G.T., Faro, A., Zhao, Y., Stickney, H., Novellasdemunt, L., Henriques, P., Gestri, G., Redhouse White, E., Ren, J., Lu, W., Young, R.M., Hawkins, T.A., Cavodeassi, F., Schwarz, Q., Dreosti, E., Raible, D.W., Li, V.S.W., Wright, G.J., Jones, E.Y., Wilson, S.W.(2024) Science 384: 573-579
- PubMed: 38696577
- DOI: https://doi.org/10.1126/science.ade6970
- Primary Citation of Related Structures:
8S7C - PubMed Abstract:
Neurons on the left and right sides of the nervous system often show asymmetric properties, but how such differences arise is poorly understood. Genetic screening in zebrafish revealed that loss of function of the transmembrane protein Cachd1 resulted in right-sided habenula neurons adopting left-sided identity. Cachd1 is expressed in neuronal progenitors, functions downstream of asymmetric environmental signals, and influences timing of the normally asymmetric patterns of neurogenesis. Biochemical and structural analyses demonstrated that Cachd1 can bind simultaneously to Lrp6 and Frizzled family Wnt co-receptors. Consistent with this, lrp6 mutant zebrafish lose asymmetry in the habenulae, and epistasis experiments support a role for Cachd1 in modulating Wnt pathway activity in the brain. These studies identify Cachd1 as a conserved Wnt receptor-interacting protein that regulates lateralized neuronal identity in the zebrafish brain.
- Cell and Developmental Biology, Division of Biosciences, University College London, London WC1E 6BT, UK.
Organizational Affiliation: