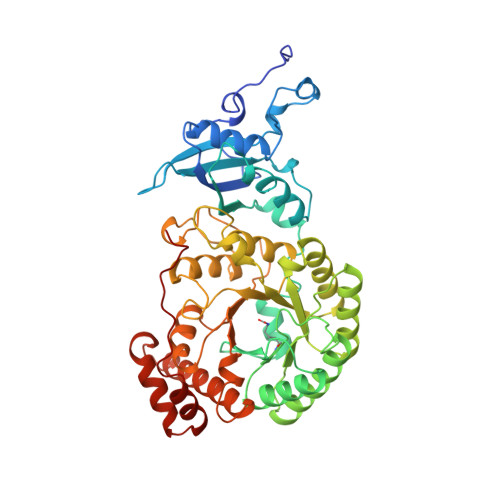
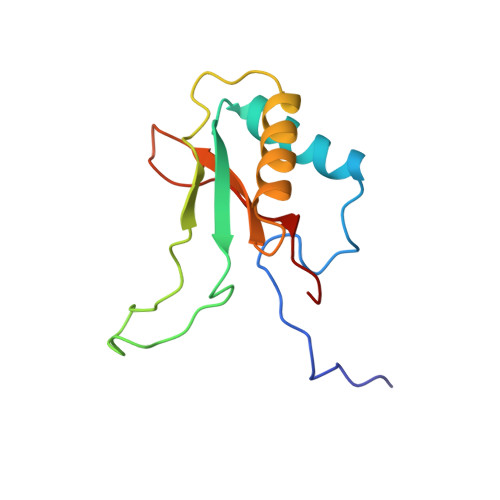
Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate.
Andersson, I.(1996) J Mol Biology 259: 160-174
- PubMed: 8648644
- DOI: https://doi.org/10.1006/jmbi.1996.0310
- Primary Citation of Related Structures:
8RUC - PubMed Abstract:
Ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from spinach is a hexadecamer (L8S8, Mr = 550,000) consisting of eight large (L, 475 residues) and eight small subunits (S, 123 residues). High-resolution data collection on crystals with large unit cells is not a trivial task due to the effect of radiation damage and the large number of overlapping reflections when conventional data collection methods are used. In order to minimise these effects, data on rubisco were collected with a giant Weissenberg camera at long crystal to image-plate distances at the synchrotron of the Photon Factory, Japan. Relative to conventional data sets, this experimental arrangement allowed a 20 to 30-fold reduction of the X-ray dose/exposure time for data collection. This paper describes the refined 1.6 A crystal structure of activated rubisco complexed with a transition state analogue, 2-carboxyarabinitol-bisphosphate. The crystallographic asymmetric unit contains an L4S4 unit, representing half of the molecule. The structure presented here is currently the highest resolution structure for any protein of comparable size. Refinement of the model was carried out by restrained least squares techniques without non-crystallographic symmetry averaging. The results show that all L and S subunits have identical three-dimensional structures, and their arrangement within the hexadecamer has no intrinsic asymmetry. A detailed analysis of the high-resolution maps identified 30 differences in the sequence of the small subunit, indicating a larger than usual heterogeneity for this nuclear encoded protein in spinach. No such differences were found in the sequence of the chloroplast encoded large subunit. The transition state analogue is in the cis conformation at the active site suggesting a key role for the carbamate of Lys201 in catalysis. Analysis of the active site around the catalytically essential magnesium ion further indicates that residues in the second liganding sphere of the metal play a role in fine-tuning the acid-base character and the position of the residues directly liganded to the metal.
- Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden.
Organizational Affiliation: