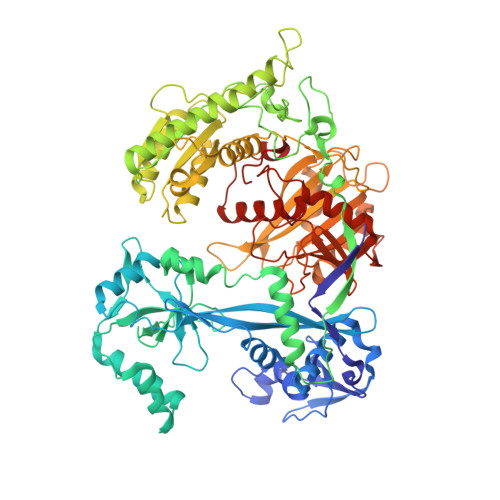
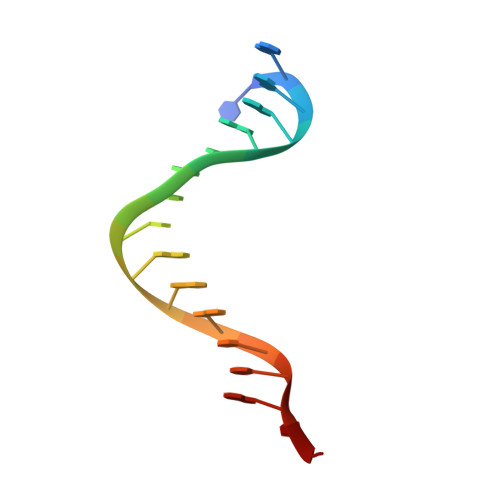
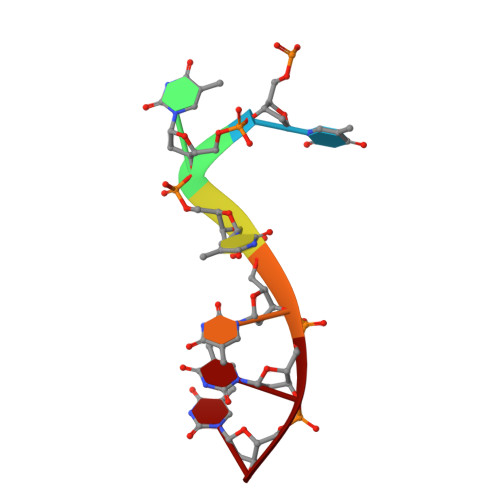
Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Wang, L., Chen, W., Zhang, C., Xie, X., Huang, F., Chen, M., Mao, W., Yu, N., Wei, Q., Ma, L., Li, Z.(2024) Mol Cell 84: 675-686.e4
- PubMed: 38295801
- DOI: https://doi.org/10.1016/j.molcel.2024.01.004
- Primary Citation of Related Structures:
8JPX, 8WD8 - PubMed Abstract:
The Argonaute nuclease from the thermophilic archaeon Pyrococcus furiosus (PfAgo) contributes to host defense and represents a promising biotechnology tool. Here, we report the structure of a PfAgo-guide DNA-target DNA ternary complex at the cleavage-compatible state. The ternary complex is predominantly dimerized, and the dimerization is solely mediated by PfAgo at PIWI-MID, PIWI-PIWI, and PAZ-N interfaces. Additionally, PfAgo accommodates a short 14-bp guide-target DNA duplex with a wedge-type N domain and specifically recognizes 5'-phosphorylated guide DNA. In contrast, the PfAgo-guide DNA binary complex is monomeric, and the engagement of target DNA with 14-bp complementarity induces sufficient dimerization and activation of PfAgo, accompanied by movement of PAZ and N domains. A closely related Argonaute from Thermococcus thioreducens adopts a similar dimerization configuration with an additional zinc finger formed at the dimerization interface. Dimerization of both Argonautes stabilizes the catalytic loops, highlighting the important role of Argonaute dimerization in the activation and target cleavage.
- State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Organizational Affiliation: