Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Wu, J., Chen, S., Wang, C., Lin, W., Huang, C., Fan, C., Han, D., Lu, D., Xu, X., Sui, S., Zhang, L.(2023) Mol Plant 16: 1937-1950
- PubMed: 37936349
- DOI: https://doi.org/10.1016/j.molp.2023.11.002
- Primary Citation of Related Structures:
8J6Z, 8J7A, 8J7B - PubMed Abstract:
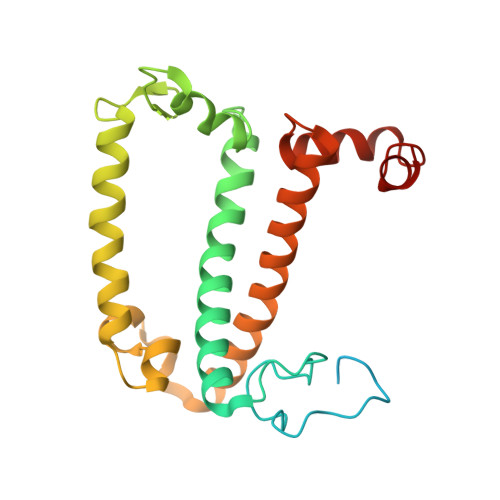
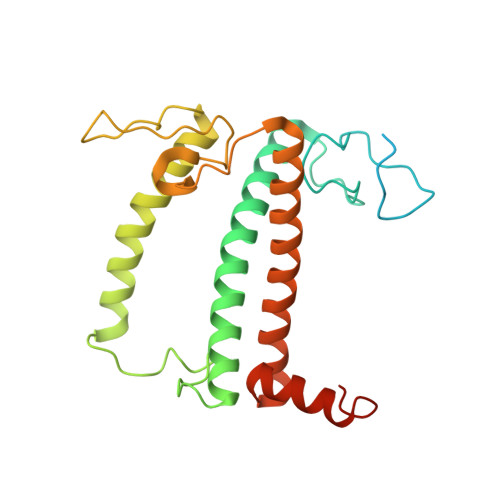
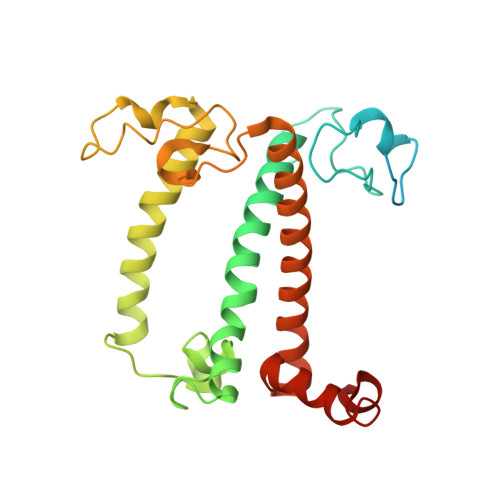
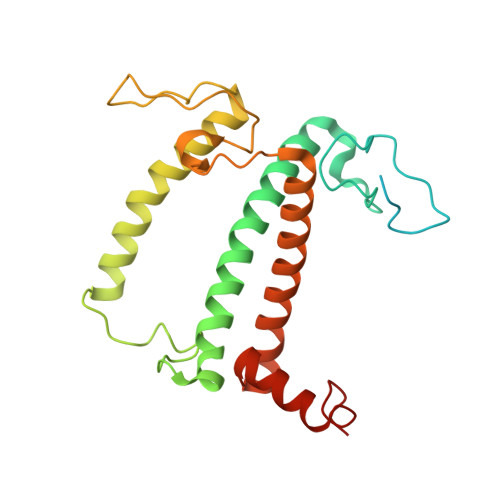
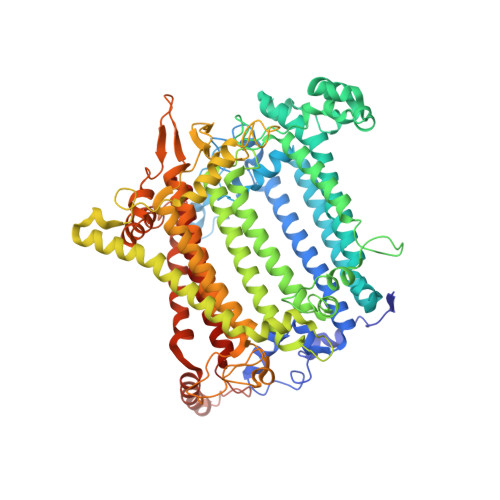
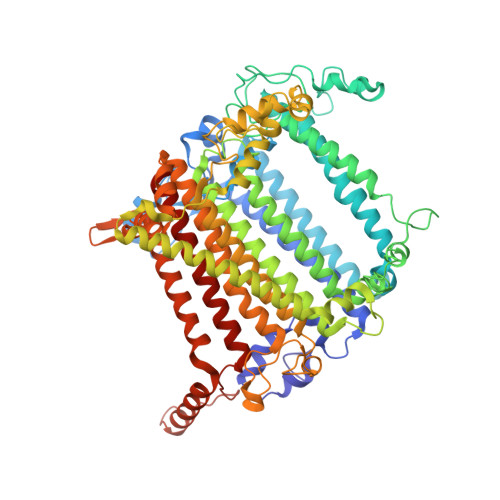
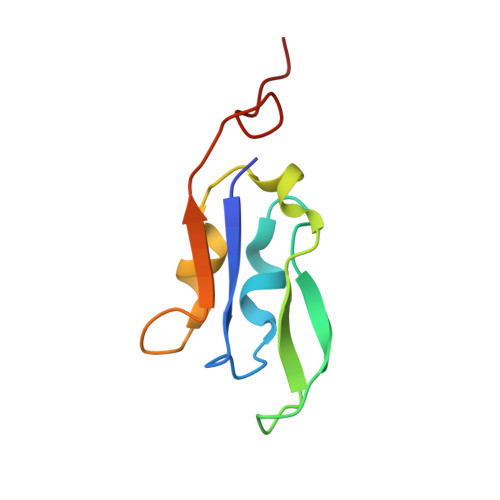
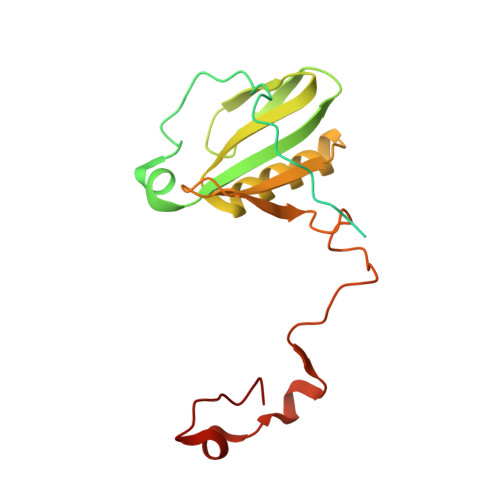
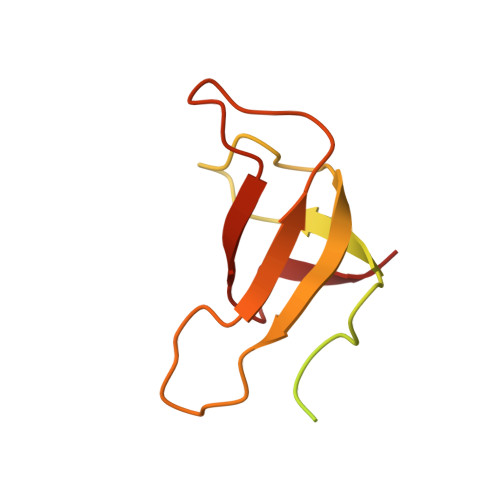
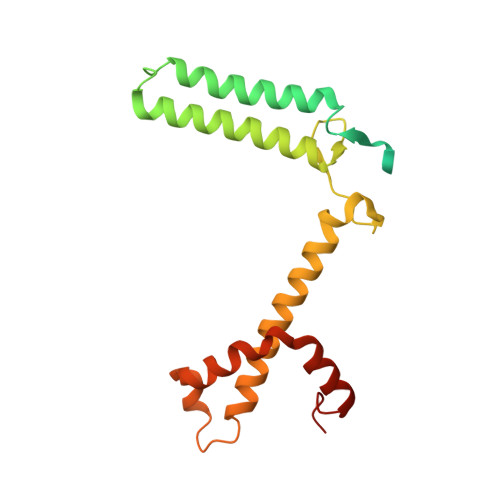
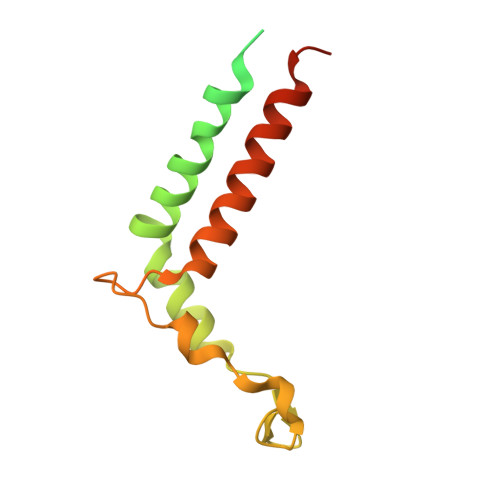
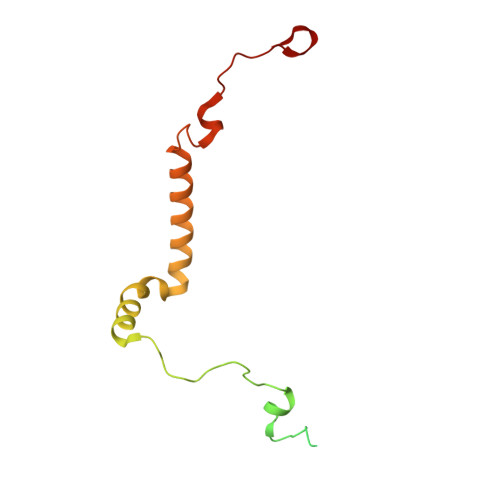
State transition is a fundamental light acclimation mechanism of photosynthetic organisms in response to the environmental light conditions. This process rebalances the excitation energy between photosystem I (PSI) and photosystem II through regulated reversible binding of the light-harvesting complex II (LHCII) to PSI. However, the structural reorganization of PSI-LHCI, the dynamic binding of LHCII, and the regulatory mechanisms underlying state transitions are less understood in higher plants. In this study, using cryoelectron microscopy we resolved the structures of PSI-LHCI in both state 1 (PSI-LHCI-ST1) and state 2 (PSI-LHCI-LHCII-ST2) from Arabidopsis thaliana. Combined genetic and functional analyses revealed novel contacts between Lhcb1 and PsaK that further enhanced the binding of the LHCII trimer to the PSI core with the known interactions between phosphorylated Lhcb2 and the PsaL/PsaH/PsaO subunits. Specifically, PsaO was absent in the PSI-LHCI-ST1 supercomplex but present in the PSI-LHCI-LHCII-ST2 supercomplex, in which the PsaL/PsaK/PsaA subunits undergo several conformational changes to strengthen the binding of PsaO in ST2. Furthermore, the PSI-LHCI module adopts a more compact configuration with shorter Mg-to-Mg distances between the chlorophylls, which may enhance the energy transfer efficiency from the peripheral antenna to the PSI core in ST2. Collectively, our work provides novel structural and functional insights into the mechanisms of light acclimation during state transitions in higher plants.
- State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Jinming Avenue, Kaifeng 475004, China.
Organizational Affiliation: