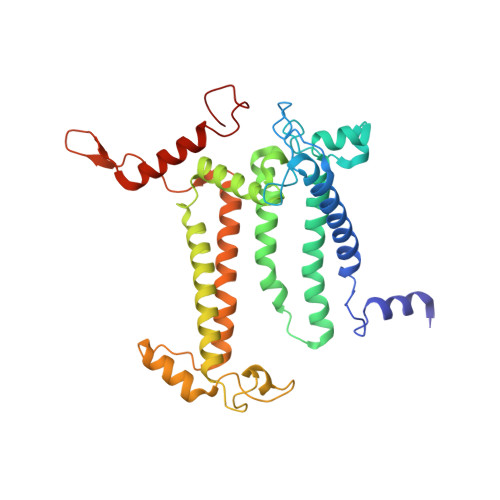
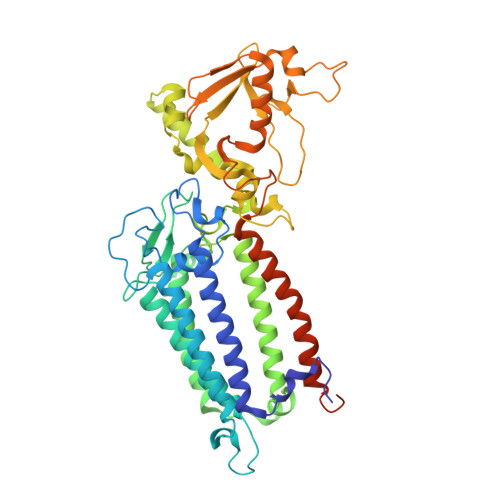
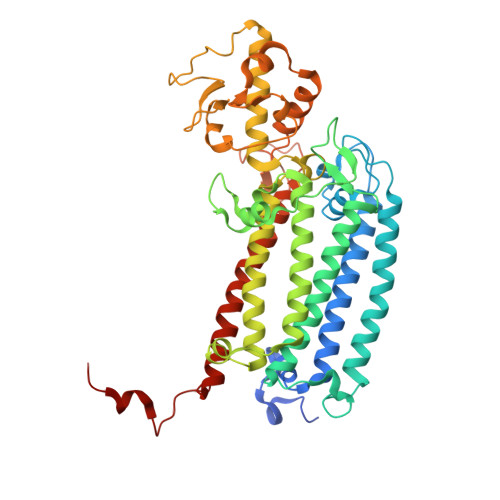
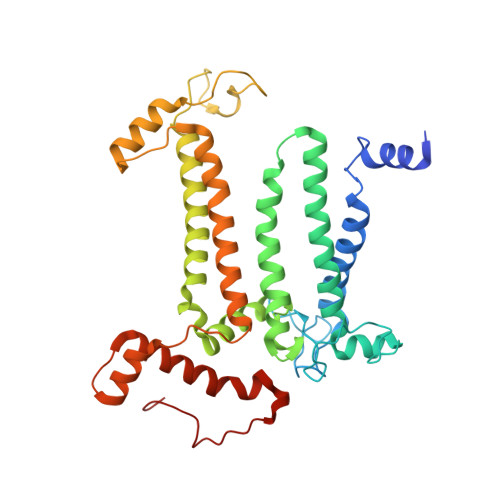
Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Feng, Y., Li, Z., Li, X., Shen, L., Liu, X., Zhou, C., Zhang, J., Sang, M., Han, G., Yang, W., Kuang, T., Wang, W., Shen, J.R.(2023) Sci Adv 9: eadi8446-eadi8446
- PubMed: 37878698
- DOI: https://doi.org/10.1126/sciadv.adi8446
- Primary Citation of Related Structures:
8IWH, 8J0D - PubMed Abstract:
Diatoms rely on fucoxanthin chlorophyll a/c -binding proteins (FCPs) for their great success in oceans, which have a great diversity in their pigment, protein compositions, and subunit organizations. We report a unique structure of photosystem II (PSII)-FCPII supercomplex from Thalassiosira pseudonana at 2.68-Å resolution by cryo-electron microscopy. FCPIIs within this PSII-FCPII supercomplex exist in dimers and monomers, and a homodimer and a heterodimer were found to bind to a PSII core. The FCPII homodimer is formed by Lhcf7 and associates with PSII through an Lhcx family antenna Lhcx6_1, whereas the heterodimer is formed by Lhcf6 and Lhcf11 and connects to the core together with an Lhcf5 monomer through Lhca2 monomer. An extended pigment network consisting of diatoxanthins, diadinoxanthins, fucoxanthins, and chlorophylls a/c is revealed, which functions in efficient light harvesting, energy transfer, and dissipation. These results provide a structural basis for revealing the energy transfer and dissipation mechanisms and also for the structural diversity of FCP antennas in diatoms.
- Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Organizational Affiliation: