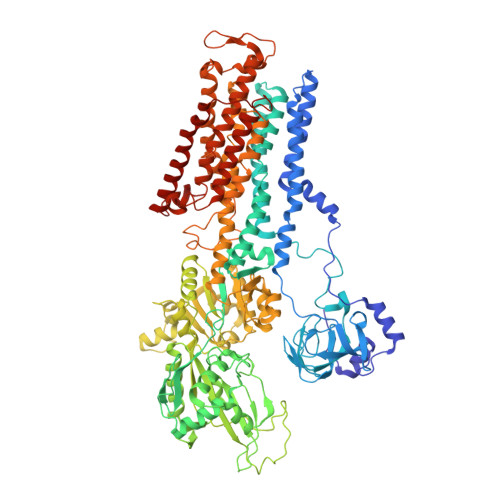
An unusual conformation from Na + -sensitive non-gastric proton pump mutants reveals molecular mechanisms of cooperative Na + -binding.
Abe, K., Nishizawa, T., Artigas, P.(2023) Biochim Biophys Acta Mol Cell Res 1870: 119543-119543
- PubMed: 37482134
- DOI: https://doi.org/10.1016/j.bbamcr.2023.119543
- Primary Citation of Related Structures:
8IJL, 8IJM - PubMed Abstract:
The Na + ,K + -ATPase (NKA) and non-gastric H + ,K + - ATPase (ngHKA) share ~65 % sequence identity, and nearly identical catalytic cycles. These pumps alternate between inward-facing (E1) and outward-facing (E2) conformations and differ in their exported substrate (Na + or H + ) and stoichiometries (3 Na + :2 K + or 1 H + :1 K + ). We reported that structures of the NKA-mimetic ngHKA mutant K794S/A797P/W940/R949C (SPWC) with 2 K + occluded in E2-P i and 3 Na + -bound in E1·ATP states were nearly identical to NKA structures in equivalent states. Here we report the cryo-EM structures of K794A and K794S, two poorly-selective ngHKA mutants, under conditions to stabilize the E1·ATP state. Unexpectedly, the structures show a hybrid with both E1- and E2-like structural features. While transmembrane segments TM1-TM3 and TM4's extracellular half adopted an E2-like conformation, the rest of the protein assumed an E1 configuration. Two spherical densities, likely bound Na + , were observed at cation-binding sites I and III, without density at site II. This explains the E2-like conformation of TM4's exoplasmic half. In NKA, oxygen atoms derived from the unwound portion of TM4 coordinated Na + at site II. Thus, the lack of Na + at site II of K794A/S prevents the luminal portion of TM4 from taking an E1-like position. The K794A structure also suggests that incomplete coordination of Na + at site III induces the halfway rotation of TM6, which impairs Na + -binding at the site II. Thus, our observations provide insight into the molecular mechanism of E2-E1 transition and cooperative Na + -binding in the NKA and other related cation pumps.
- Graduate School of Pharmaceutical Sciences, Nagoya University, Nagoya 464-8601, Japan; Cellular and Structural Physiology Institute, Nagoya University, Nagoya 464-8601, Japan; Center for One Medicine Innovative Translational Research, Gifu University Institute for Advanced Study, Japan. Electronic address: kabe@cespi.nagoya-u.ac.jp.
Organizational Affiliation: