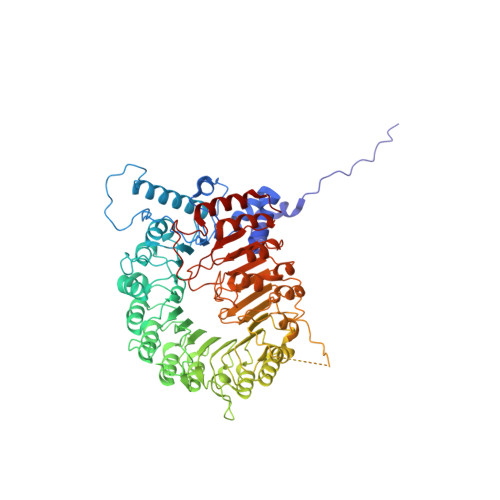
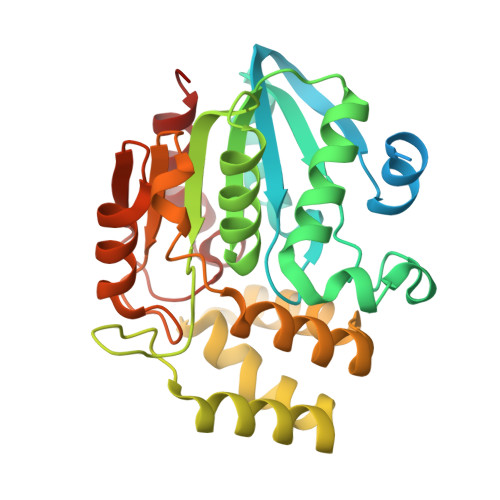
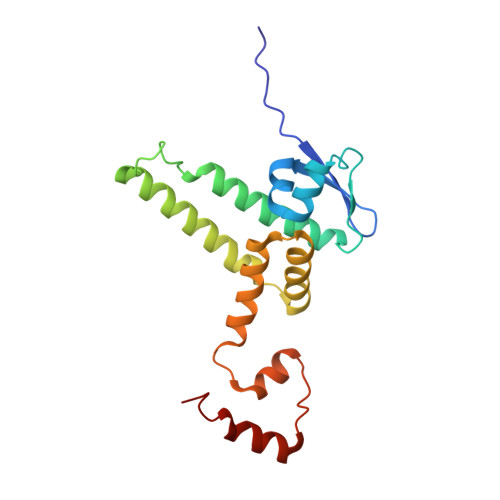
Conformational Dynamics of the D53-D3-D14 Complex in Strigolactone Signaling.
Liu, S., Wang, J., Song, B., Gong, X., Liu, H., Hu, Q., Zhang, J., Li, Q., Zheng, J., Wang, H., Xu, H.E., Li, J., Wang, B.(2023) Plant Cell Physiol 64: 1046-1056
- PubMed: 37384578
- DOI: https://doi.org/10.1093/pcp/pcad067
- Primary Citation of Related Structures:
8IF6 - PubMed Abstract:
Strigolactones (SLs) play fundamental roles in regulating plant architecture, which is a major factor determining crop yield. The perception and signal transduction of SLs require the formation of a complex containing the receptor DWARF14 (D14), an F-box protein D3 and a transcriptional regulator D53 in an SL-dependent manner. Structural and biochemical analyses of D14 and its orthologs DAD2 and AtD14, D3 and the complexes of ASK1-D3-AtD14 and D3CTH-D14 have made great contributions to understanding the mechanisms of SL perception. However, structural analyses of D53 and the D53-D3-D14 holo-complex are challenging, and the biochemical mechanism underlying the complex assembly remains poorly understood. Here, we found that apo-D53 was rather flexible and reconstituted the holo-complex containing D53, S-phase kinase-associated protein 1 (SKP1), D3 and D14 with rac-GR24. The cryo-electron microscopy (cryo-EM) structure of SKP1-D3-D14 in the presence of D53 was analyzed and superimposed on the crystal structure of ASK1-D3-AtD14 without D53. No large conformational rearrangement was observed, but a 9Å rotation appeared between D14 and AtD14. Using hydrogen-deuterium exchange monitored by mass spectrometry, we analyzed dynamic motifs of D14, D3 and D53 in the D53-SKP1-D3-D14 complex assembly process and further identified two potential interfaces in D53 that are located in the N and D2 domains, respectively. Together, our results uncovered the dynamic conformational changes and built a model of the holo-complex D53-SKP1-D3-D14, offering valuable information for the biochemical and genetic mechanisms of SL perception and signal transduction.
- Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: