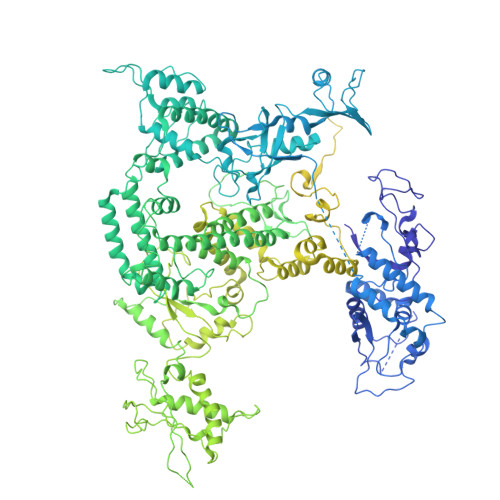
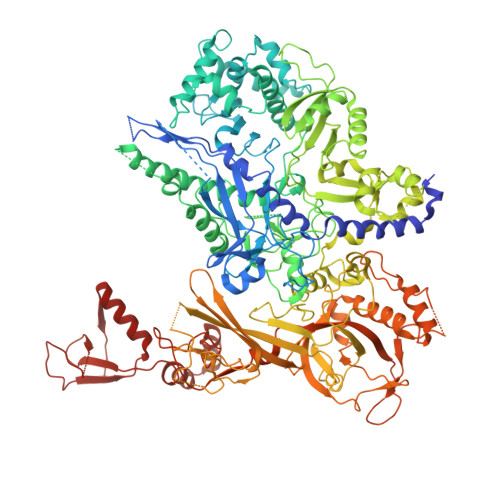
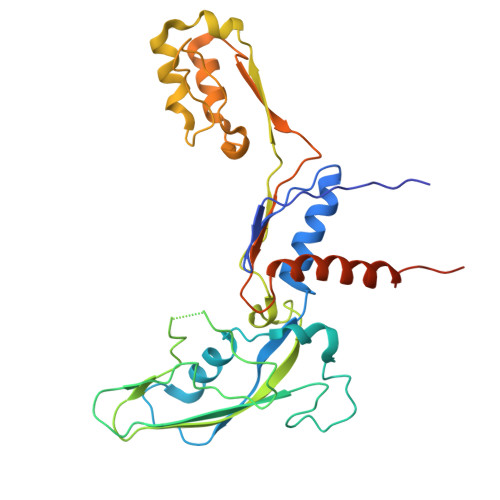
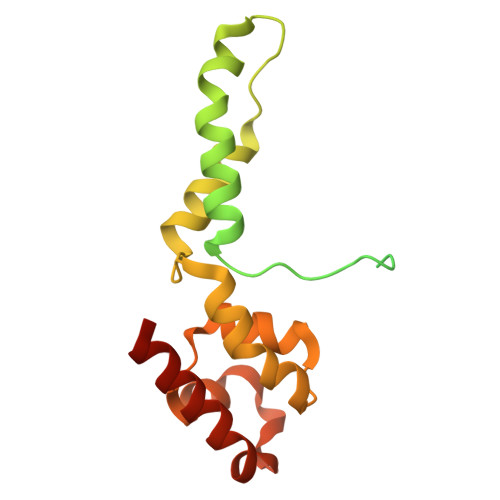
A cryo-EM structure of KTF1-bound polymerase V transcription elongation complex.
Zhang, H.W., Huang, K., Gu, Z.X., Wu, X.X., Wang, J.W., Zhang, Y.(2023) Nat Commun 14: 3118-3118
- PubMed: 37253723
- DOI: https://doi.org/10.1038/s41467-023-38619-x
- Primary Citation of Related Structures:
8HYJ - PubMed Abstract:
De novo DNA methylation in plants relies on transcription of RNA polymerase V (Pol V) along with KTF1, which produce long non-coding RNAs for recruitment and assembly of the DNA methylation machinery. Here, we report a cryo-EM structure of the Pol V transcription elongation complex bound to KTF1. The structure reveals the conformation of the structural motifs in the active site of Pol V that accounts for its inferior RNA-extension ability. The structure also reveals structural features of Pol V that prevent it from interacting with the transcription factors of Pol II and Pol IV. The KOW5 domain of KTF1 binds near the RNA exit channel of Pol V providing a scaffold for the proposed recruitment of Argonaute proteins to initiate the assembly of the DNA methylation machinery. The structure provides insight into the Pol V transcription elongation process and the role of KTF1 during Pol V transcription-coupled DNA methylation.
- Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200032, China.
Organizational Affiliation: