Structural insights into the assembly and energy transfer of the Lhcb9-dependent photosystem I from moss Physcomitrium patens.
Sun, H., Shang, H., Pan, X., Li, M.(2023) Nat Plants 9: 1347-1358
- PubMed: 37474782
- DOI: https://doi.org/10.1038/s41477-023-01463-4
- Primary Citation of Related Structures:
8HTU - PubMed Abstract:
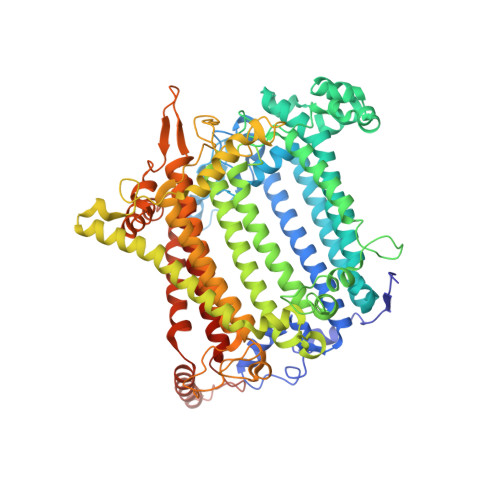
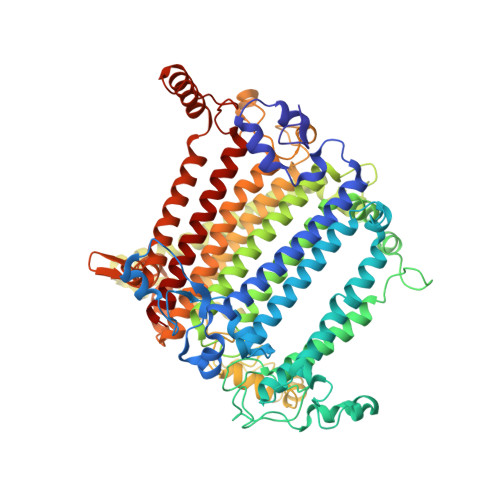
In plants and green algae, light-harvesting complexes I and II (LHCI and LHCII) constitute the antennae of photosystem I (PSI), thus effectively increasing the cross-section of the PSI core. The moss Physcomitrium patens (P. patens) represents a well-studied primary land-dwelling photosynthetic autotroph branching from the common ancestor of green algae and land plants at the early stage of evolution. P. patens possesses at least three types of PSI with different antenna sizes. The largest PSI form (PpPSI-L) exhibits a unique organization found neither in flowering plants nor in algae. Its formation is mediated by the P. patens-specific LHC protein, Lhcb9. While previous studies have revealed the overall architecture of PpPSI-L, its assembly details and the relationship between different PpPSI types remain unclear. Here we report the high-resolution structure of PpPSI-L. We identified 14 PSI core subunits, one Lhcb9, one phosphorylated LHCII trimer and eight LHCI monomers arranged as two belts. Our structural analysis established the essential role of Lhcb9 and the phosphorylated LHCII in stabilizing the complex. In addition, our results suggest that PpPSI switches between different types, which share identical modules. This feature may contribute to the dynamic adjustment of the light-harvesting capability of PSI under different light conditions.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: