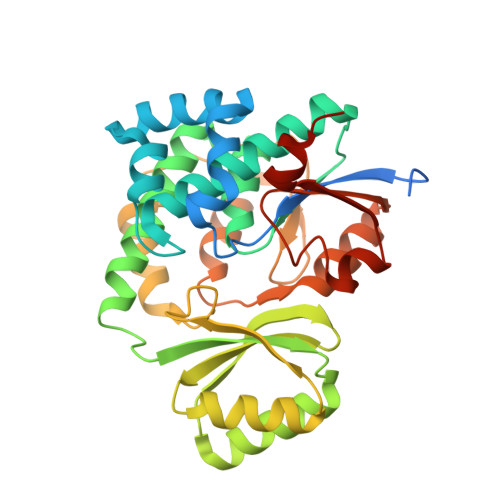
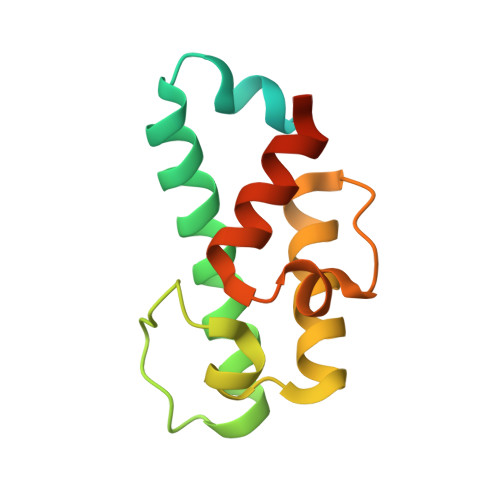
Structural Basis of Transient Interactions of Acyltransferase VinK with the Loading Acyl Carrier Protein of the Vicenistatin Modular Polyketide Synthase.
Miyanaga, A., Kawada, K., Chisuga, T., Kudo, F., Eguchi, T.(2023) Biochemistry 62: 17-21
- PubMed: 36512613
- DOI: https://doi.org/10.1021/acs.biochem.2c00645
- Primary Citation of Related Structures:
8H6S - PubMed Abstract:
Acyltransferase (AT) recognizes its cognate acyl carrier protein (ACP) for functional transfer of an acyl unit in polyketide biosynthesis. However, structural characterization of AT-ACP complexes is limited because of the weak and transient interactions between them. In the biosynthesis of macrolactam polyketide vicenistatin, the trans -acting loading AT VinK transfers a dipeptidyl unit from the stand-alone ACP VinL to the ACP domain (VinP1ACP L ) of the loading module of modular polyketide synthase VinP1. Although the previously determined structure of the VinK-VinL complex clearly illustrates the VinL recognition mechanism of VinK, how VinK recognizes VinP1ACP L remains unclear. Here, the crystal structure of a covalent VinK-VinP1ACP L complex formed with a pantetheine-type cross-linking probe is reported at 3.0 Å resolution. The structure of the VinK-VinP1ACP L complex provides detailed insights into the transient interactions between VinK and VinP1ACP L . The importance of residues in the binding interface was confirmed by site-directed mutational analyses. The binding interface between VinK and VinP1ACP L is similar to that between VinK and VinL, although some of the interface residues are different. However, the ACP orientation and interaction mode observed in the VinK-VinP1ACP L complex are different from those observed in other AT-ACP complexes such as the disorazole trans -AT-ACP complex and cis -AT-ACP complexes of modular polyketide synthases. Thus, AT-ACP binding interface interactions are different in each type of AT-ACP pair.
- Department of Chemistry, Tokyo Institute of Technology, 2-12-1 O-okayama, Meguro-ku, Tokyo 152-8551, Japan.
Organizational Affiliation: