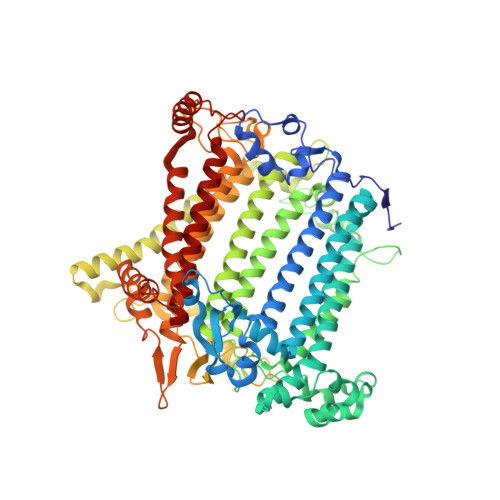
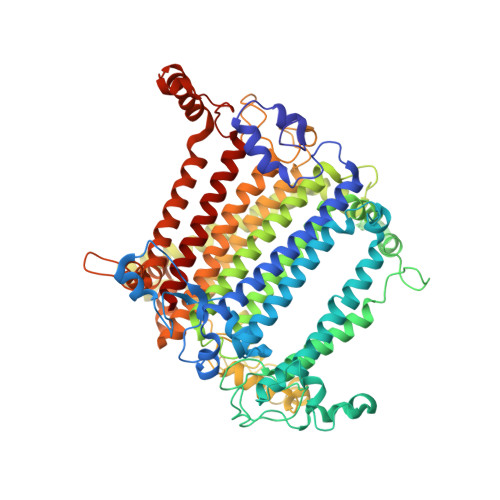
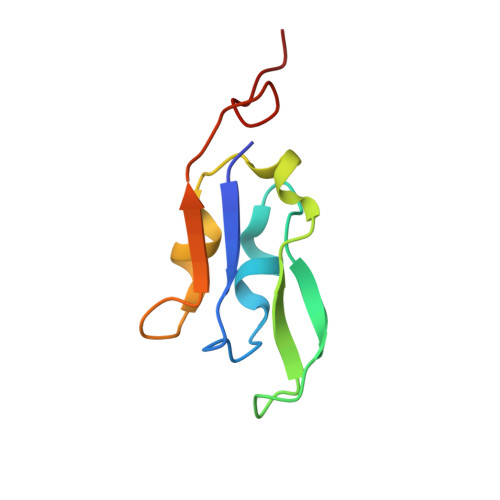
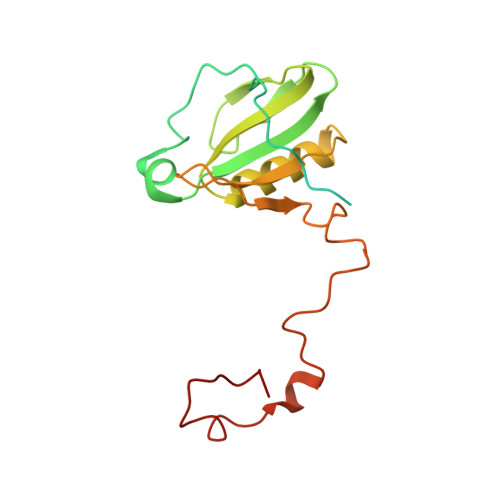
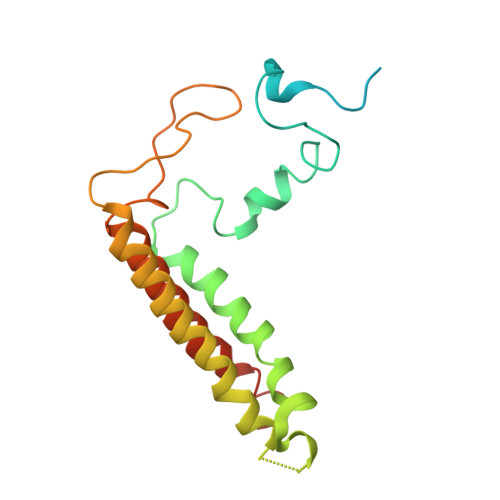
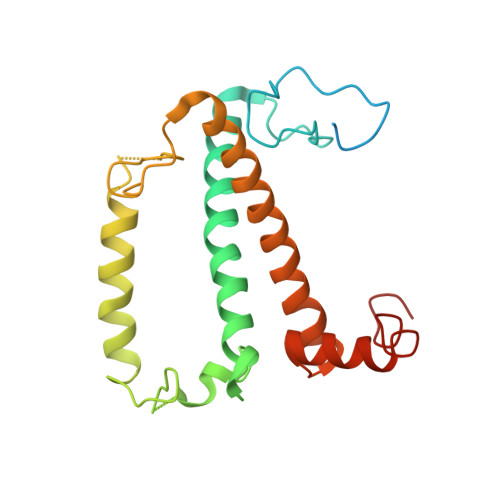
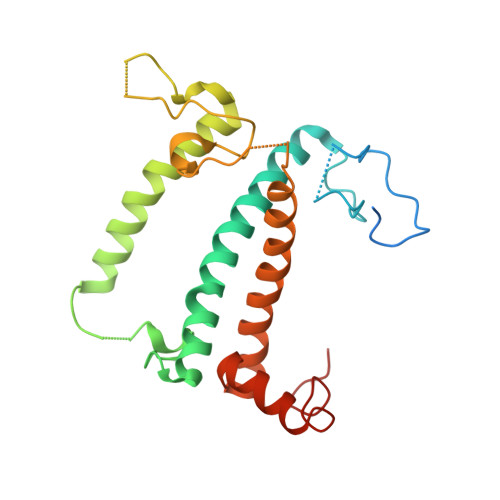
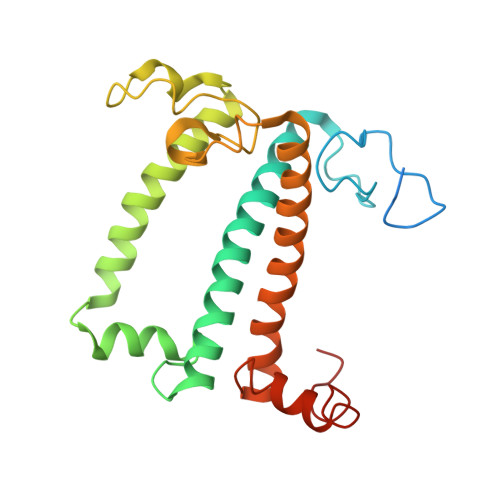
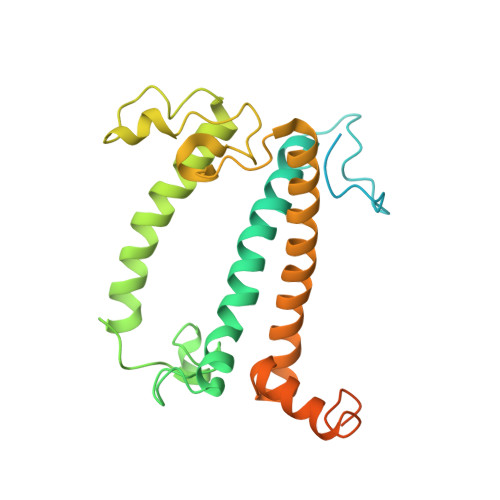
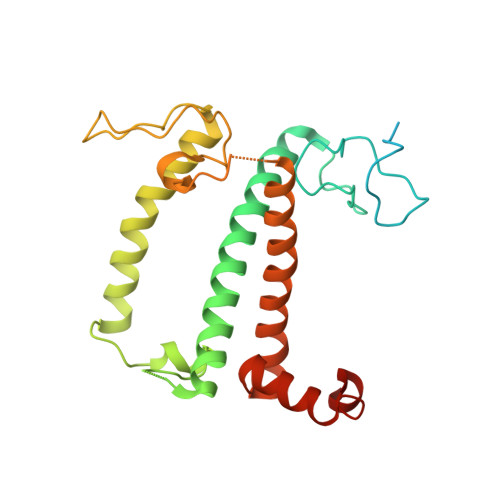
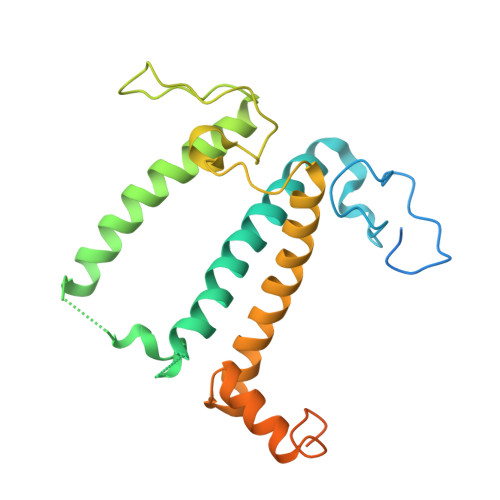
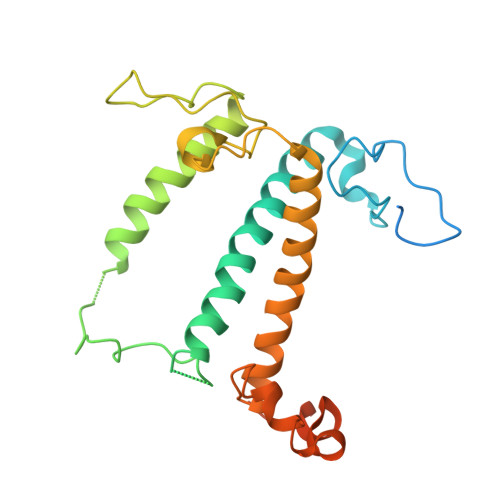
Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Gerle, C., Misumi, Y., Kawamoto, A., Tanaka, H., Kubota-Kawai, H., Tokutsu, R., Kim, E., Chorev, D., Abe, K., Robinson, C.V., Mitsuoka, K., Minagawa, J., Kurisu, G.(2023) Biochim Biophys Acta Bioenerg 1864: 148986-148986
- PubMed: 37270022
- DOI: https://doi.org/10.1016/j.bbabio.2023.148986
- Primary Citation of Related Structures:
7WYI, 7WZN, 8H2U - PubMed Abstract:
Photosystem I (PSI) from the green alga Chlamydomonas reinhardtii, with various numbers of membrane bound antenna complexes (LHCI), has been described in great detail. In contrast, structural characterization of soluble binding partners is less advanced. Here, we used X-ray crystallography and single particle cryo-EM to investigate three structures of the PSI-LHCI supercomplex from Chlamydomonas reinhardtii. An X-ray structure demonstrates the absence of six chlorophylls from the luminal side of the LHCI belts, suggesting these pigments were either physically absent or less stably associated with the complex, potentially influencing excitation transfer significantly. CryoEM revealed extra densities on luminal and stromal sides of the supercomplex, situated in the vicinity of the electron transfer sites. These densities disappeared after the binding of oxidized ferredoxin to PSI-LHCI. Based on these structures, we propose the existence of a PSI-LHCI resting state with a reduced active chlorophyll content, electron donors docked in waiting positions and regulatory binding partners positioned at the electron acceptor site. The resting state PSI-LHCI supercomplex would be recruited to its active form by the availability of oxidized ferredoxin.
- Life Science Research Infrastructure Group, RIKEN SPring-8 Center, Kouto, Hyogo, Japan; Laboratory for Protein Crystallography, Institute for Protein Research, Osaka University, Suita, Osaka, Japan. Electronic address: christoph.gerle@riken.jp.
Organizational Affiliation: