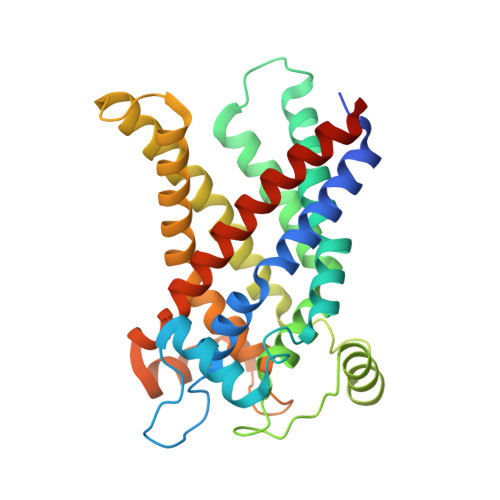
Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Jones, S.A., Gogoi, P., Ruprecht, J.J., King, M.S., Lee, Y., Zogg, T., Pardon, E., Chand, D., Steimle, S., Copeman, D.M., Cotrim, C.A., Steyaert, J., Crichton, P.G., Moiseenkova-Bell, V., Kunji, E.R.S.(2023) Sci Adv 9: eadh4251-eadh4251
- PubMed: 37256948
- DOI: https://doi.org/10.1126/sciadv.adh4251
- Primary Citation of Related Structures:
8G8W - PubMed Abstract:
Mitochondrial uncoupling protein 1 (UCP1) gives brown adipose tissue of mammals its specialized ability to burn calories as heat for thermoregulation. When activated by fatty acids, UCP1 catalyzes the leak of protons across the mitochondrial inner membrane, short-circuiting the mitochondrion to generate heat, bypassing ATP synthesis. In contrast, purine nucleotides bind and inhibit UCP1, regulating proton leak by a molecular mechanism that is unclear. We present the cryo-electron microscopy structure of the GTP-inhibited state of UCP1, which is consistent with its nonconducting state. The purine nucleotide cross-links the transmembrane helices of UCP1 with an extensive interaction network. Our results provide a structural basis for understanding the specificity and pH dependency of the regulatory mechanism. UCP1 has retained all of the key functional and structural features required for a mitochondrial carrier-like transport mechanism. The analysis shows that inhibitor binding prevents the conformational changes that UCP1 uses to facilitate proton leak.
- MRC Mitochondrial Biology Unit, University of Cambridge, Cambridge Biomedical Campus, Keith Peters Building, Cambridge CB2 0XY, UK.
Organizational Affiliation: