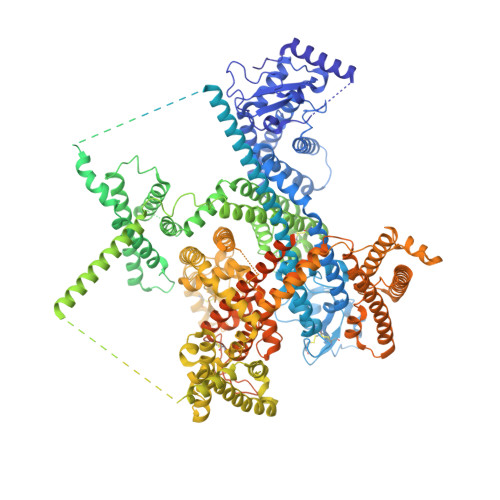
Cannabidiol inhibits Na v channels through two distinct binding sites.
Huang, J., Fan, X., Jin, X., Jo, S., Zhang, H.B., Fujita, A., Bean, B.P., Yan, N.(2023) Nat Commun 14: 3613-3613
- PubMed: 37330538
- DOI: https://doi.org/10.1038/s41467-023-39307-6
- Primary Citation of Related Structures:
8G1A - PubMed Abstract:
Cannabidiol (CBD), a major non-psychoactive phytocannabinoid in cannabis, is an effective treatment for some forms of epilepsy and pain. At high concentrations, CBD interacts with a huge variety of proteins, but which targets are most relevant for clinical actions is still unclear. Here we show that CBD interacts with Na v 1.7 channels at sub-micromolar concentrations in a state-dependent manner. Electrophysiological experiments show that CBD binds to the inactivated state of Na v 1.7 channels with a dissociation constant of about 50 nM. The cryo-EM structure of CBD bound to Na v 1.7 channels reveals two distinct binding sites. One is in the IV-I fenestration near the upper pore. The other binding site is directly next to the inactivated "wedged" position of the Ile/Phe/Met (IFM) motif on the short linker between repeats III and IV, which mediates fast inactivation. Consistent with producing a direct stabilization of the inactivated state, mutating residues in this binding site greatly reduced state-dependent binding of CBD. The identification of this binding site may enable design of compounds with improved properties compared to CBD itself.
- Department of Molecular Biology, Princeton University, Princeton, NJ, 08544, USA.
Organizational Affiliation: