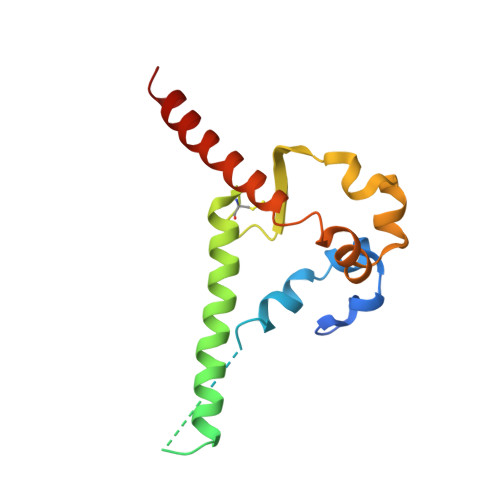
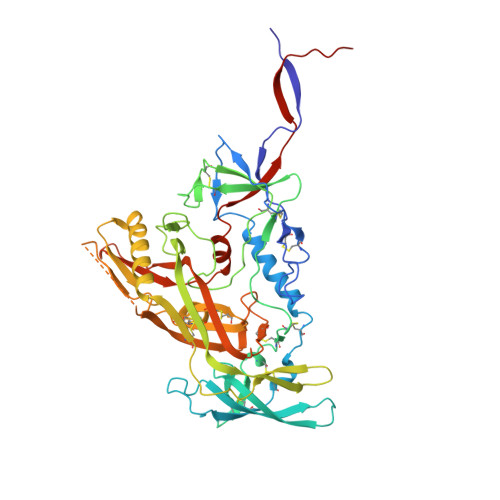
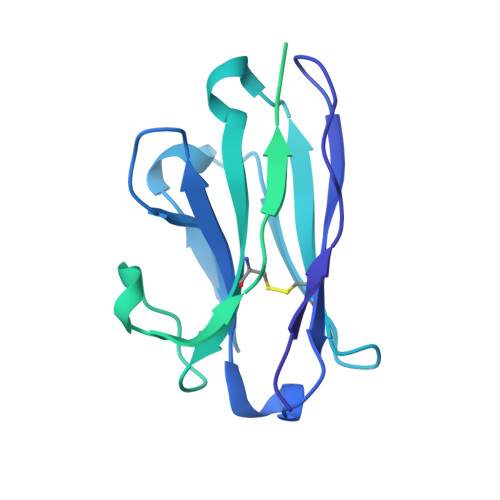
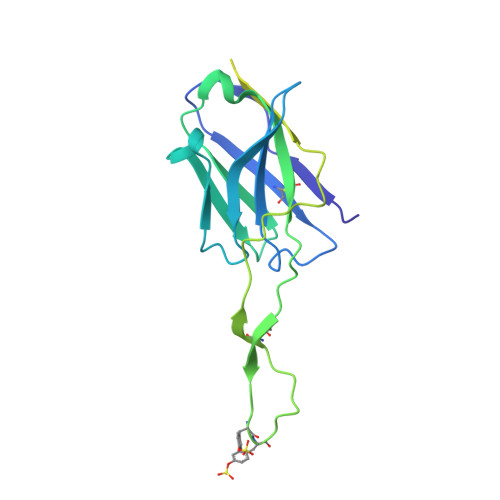
Bispecific antibody CAP256.J3LS targets V2-apex and CD4-binding sites with high breadth and potency.
Zhang, B., Gorman, J., Kwon, Y.D., Pegu, A., Chao, C.W., Liu, T., Asokan, M., Bender, M.F., Bylund, T., Damron, L., Gollapudi, D., Lei, P., Li, Y., Liu, C., Louder, M.K., McKee, K., Olia, A.S., Rawi, R., Schon, A., Wang, S., Yang, E.S., Yang, Y., Carlton, K., Doria-Rose, N.A., Shapiro, L., Seaman, M.S., Mascola, J.R., Kwong, P.D.(2023) MAbs 15: 2165390-2165390
- PubMed: 36729903
- DOI: https://doi.org/10.1080/19420862.2023.2165390
- Primary Citation of Related Structures:
8FIS - PubMed Abstract:
Antibody CAP256-VRC26.25 targets the second hypervariable region (V2) at the apex of the HIV envelope (Env) trimer with extraordinary neutralization potency, although less than optimal breadth. To improve breadth, we linked the light chain of CAP256V2LS, an optimized version of CAP256-VRC26.25 currently under clinical evaluation, to the llama nanobody J3, which has broad CD4-binding site-directed neutralization. The J3-linked bispecific antibody exhibited improved breadth and potency over both J3 and CAP256V2LS, indicative of synergistic neutralization. The cryo-EM structure of the bispecific antibody in complex with a prefusion-closed Env trimer revealed simultaneous binding of J3 and CAP256V2LS. We further optimized the pharmacokinetics of the bispecific antibody by reducing the net positive charge of J3. The optimized bispecific antibody, which we named CAP256.J3LS, had a half-life similar to CAP256V2LS in human FcRn knock-in mice and exhibited suitable auto-reactivity, manufacturability, and biophysical risk. CAP256.J3LS neutralized over 97% of a multiclade 208-strain panel (geometric mean concentration for 80% inhibition (IC 80 ) 0.079 μg/ml) and 100% of a 100-virus clade C panel (geometric mean IC 80 of 0.05 μg/ml), suggesting its anti-HIV utility especially in regions where clade C dominates.
- Vaccine Research Center, NIAID, National Institutes of Health, Bethesda, MD, USA.
Organizational Affiliation: