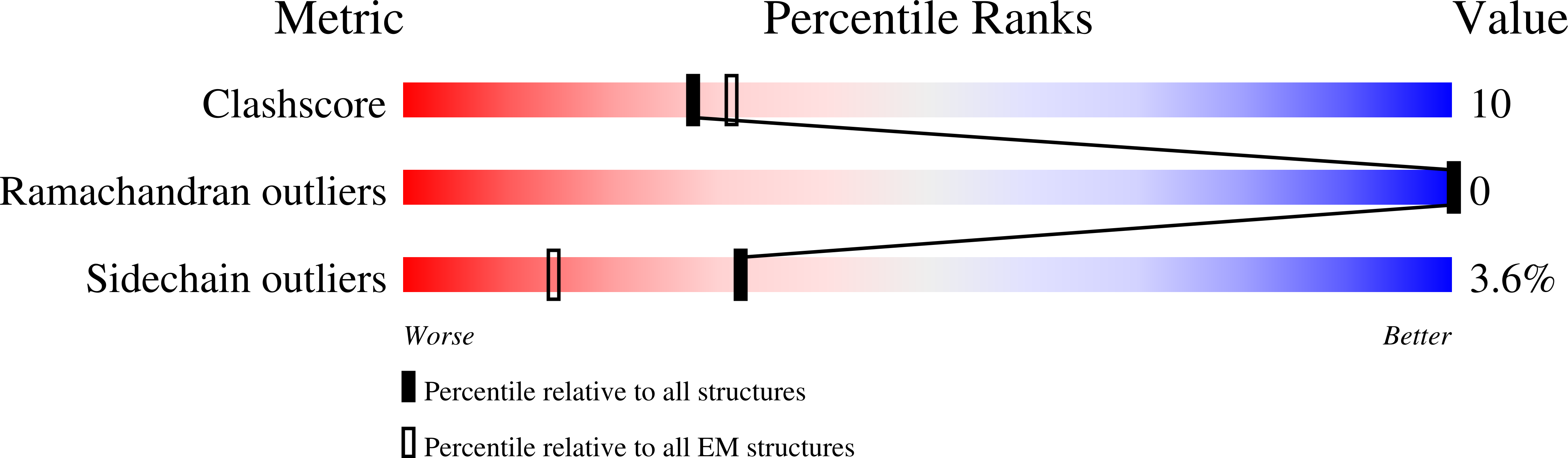Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Fan, X., Huang, J., Jin, X., Yan, N.(2023) Proc Natl Acad Sci U S A 120: e2220578120-e2220578120
- PubMed: 36696443
- DOI: https://doi.org/10.1073/pnas.2220578120
- Primary Citation of Related Structures:
8FHD - PubMed Abstract:
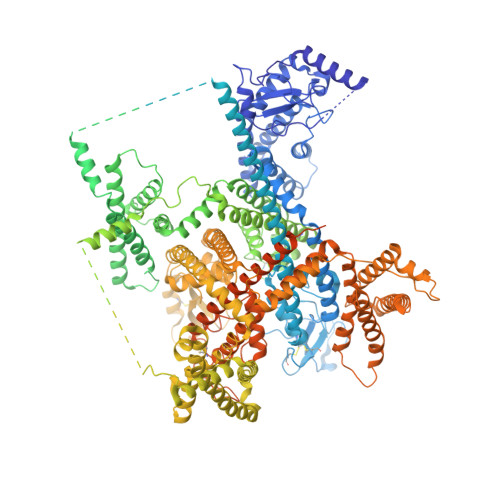
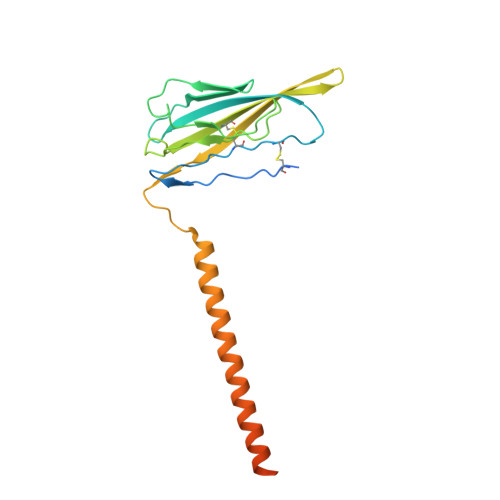
Voltage-gated sodium channel Na v 1.6 plays a crucial role in neuronal firing in the central nervous system (CNS). Aberrant function of Na v 1.6 may lead to epilepsy and other neurological disorders. Specific inhibitors of Na v 1.6 thus have therapeutic potentials. Here we present the cryo-EM structure of human Na v 1.6 in the presence of auxiliary subunits β1 and fibroblast growth factor homologous factor 2B (FHF2B) at an overall resolution of 3.1 Å. The overall structure represents an inactivated state with closed pore domain (PD) and all "up" voltage-sensing domains. A conserved carbohydrate-aromatic interaction involving Trp302 and Asn326, together with the β1 subunit, stabilizes the extracellular loop in repeat I. Apart from regular lipids that are resolved in the EM map, an unprecedented Y-shaped density that belongs to an unidentified molecule binds to the PD, revealing a potential site for developing Na v 1.6-specific blockers. Structural mapping of disease-related Na v 1.6 mutations provides insights into their pathogenic mechanism.
- Department of Molecular Biology, Princeton University, Princeton, NJ 08544.
Organizational Affiliation: