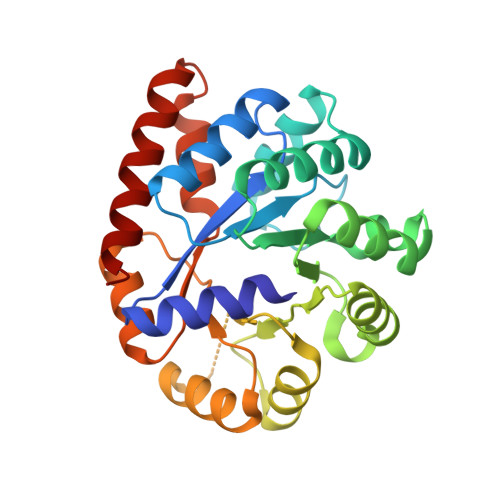
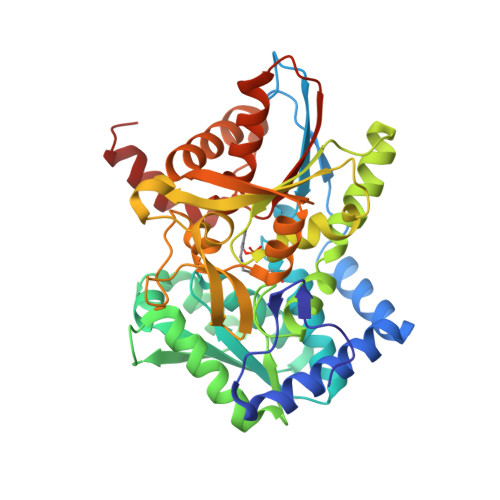
Neutron diffraction from a microgravity-grown crystal reveals the active site hydrogens of the internal aldimine form of tryptophan synthase.
Drago, V.N., Devos, J.M., Blakeley, M.P., Forsyth, V.T., Parks, J.M., Kovalevsky, A., Mueser, T.C.(2024) Cell Rep Phys Sci 5
- PubMed: 38645802
- DOI: https://doi.org/10.1016/j.xcrp.2024.101827
- Primary Citation of Related Structures:
8EYP, 8EYS, 8EZC - PubMed Abstract:
Pyridoxal 5'-phosphate (PLP), the biologically active form of vitamin B 6 , is an essential cofactor in many biosynthetic pathways. The emergence of PLP-dependent enzymes as drug targets and biocatalysts, such as tryptophan synthase (TS), has underlined the demand to understand PLP-dependent catalysis and reaction specificity. The ability of neutron diffraction to resolve the positions of hydrogen atoms makes it an ideal technique to understand how the electrostatic environment and selective protonation of PLP regulates PLP-dependent activities. Facilitated by microgravity crystallization of TS with the Toledo Crystallization Box, we report the 2.1 Å joint X-ray/neutron (XN) structure of TS with PLP in the internal aldimine form. Positions of hydrogens were directly determined in both the α- and β-active sites, including PLP cofactor. The joint XN structure thus provides insight into the selective protonation of the internal aldimine and the electrostatic environment of TS necessary to understand the overall catalytic mechanism.
- Department of Chemistry and Biochemistry, University of Toledo, Toledo, OH 43606, USA.
Organizational Affiliation: