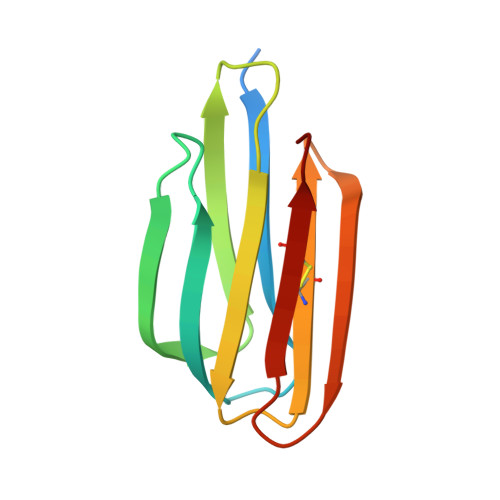
The structure of Mycobacterium thermoresistibile MmpS5 reveals a conserved disulfide bond across mycobacteria.
Cuthbert, B.J., Mendoza, J., de Miranda, R., Papavinasasundaram, K., Sassetti, C.M., Goulding, C.W.(2024) Metallomics 16
- PubMed: 38425033
- DOI: https://doi.org/10.1093/mtomcs/mfae011
- Primary Citation of Related Structures:
8EM5 - PubMed Abstract:
The tuberculosis (TB) emergency has been a pressing health threat for decades. With the emergence of drug-resistant TB and complications from the COVID-19 pandemic, the TB health crisis is more serious than ever. Mycobacterium tuberculosis (Mtb), the causative agent of TB, requires iron for its survival. Thus, Mtb has evolved several mechanisms to acquire iron from the host. Mtb produces two siderophores, mycobactin and carboxymycobactin, which scavenge for host iron. Mtb siderophore-dependent iron acquisition requires the export of apo-siderophores from the cytosol to the host environment and import of iron-bound siderophores. The export of Mtb apo-siderophores across the inner membrane is facilitated by two mycobacterial inner membrane proteins with their cognate periplasmic accessory proteins, designated MmpL4/MmpS4 and MmpL5/MmpS5. Notably, the Mtb MmpL4/MmpS4 and MmpL5/MmpS5 complexes have also been implicated in the efflux of anti-TB drugs. Herein, we solved the crystal structure of M. thermoresistibile MmpS5. The MmpS5 structure reveals a previously uncharacterized, biologically relevant disulfide bond that appears to be conserved across the Mycobacterium MmpS4/S5 homologs, and comparison with structural homologs suggests that MmpS5 may be dimeric.
- Department of Molecular Biology & Biochemistry, University of California Irvine, Irvine, CA 92697, USA.
Organizational Affiliation: