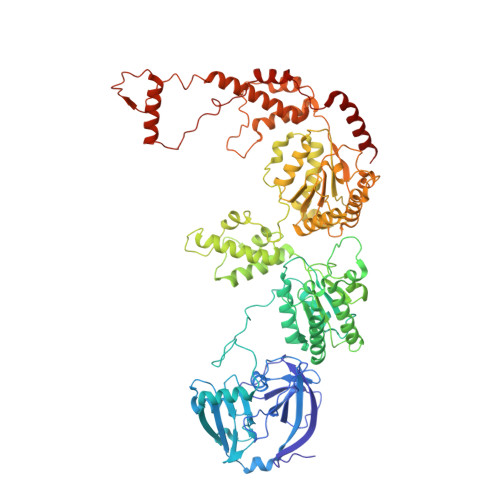
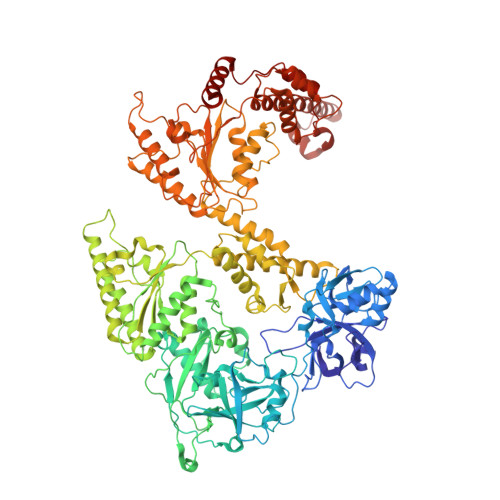
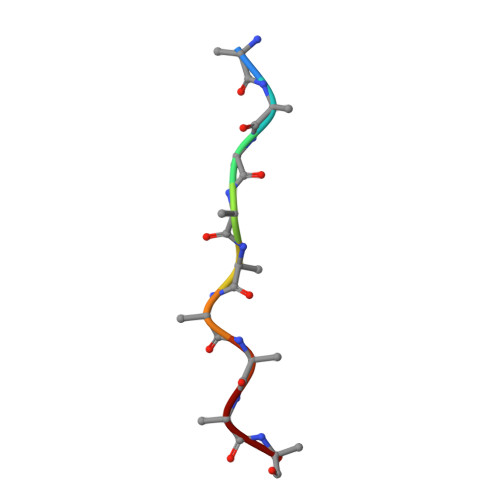
Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Ruttermann, M., Koci, M., Lill, P., Geladas, E.D., Kaschani, F., Klink, B.U., Erdmann, R., Gatsogiannis, C.(2023) Nat Commun 14: 5942-5942
- PubMed: 37741838
- DOI: https://doi.org/10.1038/s41467-023-41640-9
- Primary Citation of Related Structures:
8C0V, 8C0W - PubMed Abstract:
The double-ring AAA+ ATPase Pex1/Pex6 is required for peroxisomal receptor recycling and is essential for peroxisome formation. Pex1/Pex6 mutations cause severe peroxisome associated developmental disorders. Despite its pathophysiological importance, mechanistic details of the heterohexamer are not yet available. Here, we report cryoEM structures of Pex1/Pex6 from Saccharomyces cerevisiae, with an endogenous protein substrate trapped in the central pore of the catalytically active second ring (D2). Pairs of Pex1/Pex6(D2) subdomains engage the substrate via a staircase of pore-1 loops with distinct properties. The first ring (D1) is catalytically inactive but undergoes significant conformational changes resulting in alternate widening and narrowing of its pore. These events are fueled by ATP hydrolysis in the D2 ring and disengagement of a "twin-seam" Pex1/Pex6(D2) heterodimer from the staircase. Mechanical forces are propagated in a unique manner along Pex1/Pex6 interfaces that are not available in homo-oligomeric AAA-ATPases. Our structural analysis reveals the mechanisms of how Pex1 and Pex6 coordinate to achieve substrate translocation.
- Institute for Medical Physics and Biophysics, University Münster, Münster, Germany.
Organizational Affiliation: