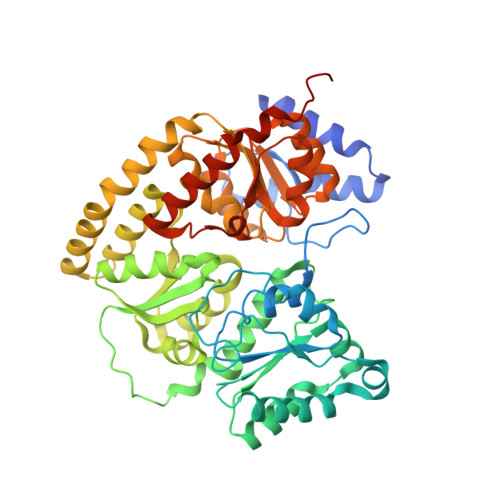
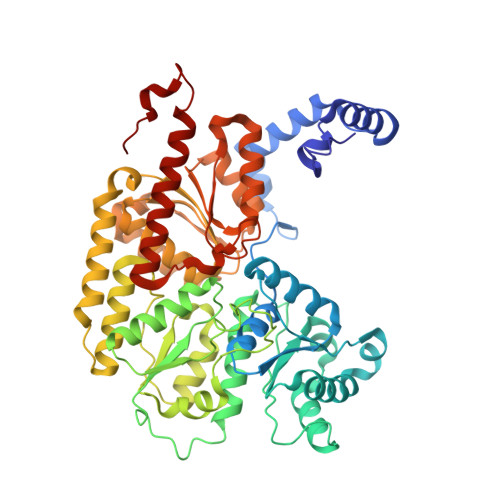
Nitrogen Fixation and Hydrogen Evolution by Sterically Encumbered Mo-Nitrogenase.
Cadoux, C., Ratcliff, D., Maslac, N., Gu, W., Tsakoumagkos, I., Hoogendoorn, S., Wagner, T., Milton, R.D.(2023) JACS Au 3: 1521-1533
- PubMed: 37234119
- DOI: https://doi.org/10.1021/jacsau.3c00165
- Primary Citation of Related Structures:
8BTS - PubMed Abstract:
The substrate-reducing proteins of all nitrogenases (MoFe, VFe, and FeFe) are organized as α 2 ß 2 (γ 2 ) multimers with two functional halves. While their dimeric organization could afford improved structural stability of nitrogenases in vivo , previous research has proposed both negative and positive cooperativity contributions with respect to enzymatic activity. Here, a 1.4 kDa peptide was covalently introduced in the proximity of the P cluster, corresponding to the Fe protein docking position. The Strep-tag carried by the added peptide simultaneously sterically inhibits electron delivery to the MoFe protein and allows the isolation of partially inhibited MoFe proteins (where the half-inhibited MoFe protein was targeted). We confirm that the partially functional MoFe protein retains its ability to reduce N 2 to NH 3 , with no significant difference in selectivity over obligatory/parasitic H 2 formation. Our experiment concludes that wild-type nitrogenase exhibits negative cooperativity during the steady state regarding H 2 and NH 3 formation (under Ar or N 2 ), with one-half of the MoFe protein inhibiting turnover in the second half. This emphasizes the presence and importance of long-range (>95 Å) protein-protein communication in biological N 2 fixation in Azotobacter vinelandii .
- Department of Inorganic and Analytical Chemistry, Faculty of Sciences, University of Geneva, Quai Ernest-Ansermet 30, 1211 Geneva 4, Switzerland.
Organizational Affiliation: