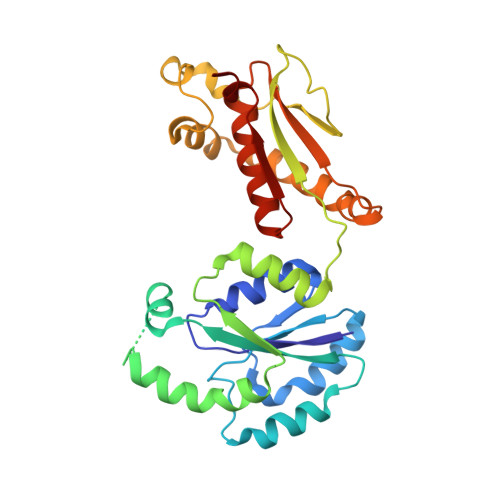
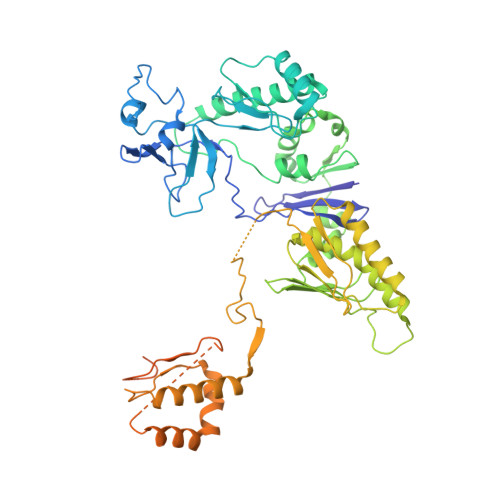
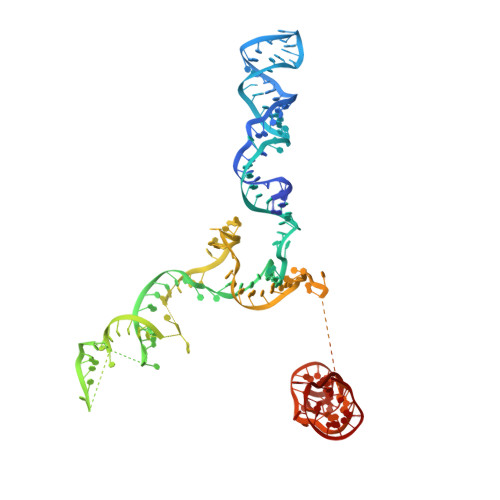
Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Islam, M.S., Hardwick, S.W., Quell, L., Durica-Mitic, S., Chirgadze, D.Y., Gorke, B., Luisi, B.F.(2023) EMBO J 42: e112574-e112574
- PubMed: 36504162
- DOI: https://doi.org/10.15252/embj.2022112574
- Primary Citation of Related Structures:
8B0I, 8B0J - PubMed Abstract:
Biogenesis of the essential precursor of the bacterial cell envelope, glucosamine-6-phosphate (GlcN6P), is controlled by intricate post-transcriptional networks mediated by GlmZ, a small regulatory RNA (sRNA). GlmZ stimulates translation of the mRNA encoding GlcN6P synthtase in Escherichia coli, but when bound by RapZ protein, the sRNA becomes inactivated through cleavage by the endoribonuclease RNase E. Here, we report the cryoEM structure of the RapZ:GlmZ complex, revealing a complementary match of the RapZ tetrameric quaternary structure to structural repeats in the sRNA. The nucleic acid is contacted by RapZ mostly through a highly conserved domain that shares an evolutionary relationship with phosphofructokinase and suggests links between metabolism and riboregulation. We also present the structure of a precleavage intermediate formed between the binary RapZ:GlmZ complex and RNase E that reveals how GlmZ is presented and recognised by the enzyme. The structures provide a framework for understanding how other encounter complexes might guide recognition and action of endoribonucleases on target transcripts, and how structured substrates in polycistronic precursors may be recognised for processing by RNase E.
- Department of Biochemistry, University of Cambridge, Cambridge, UK.
Organizational Affiliation: