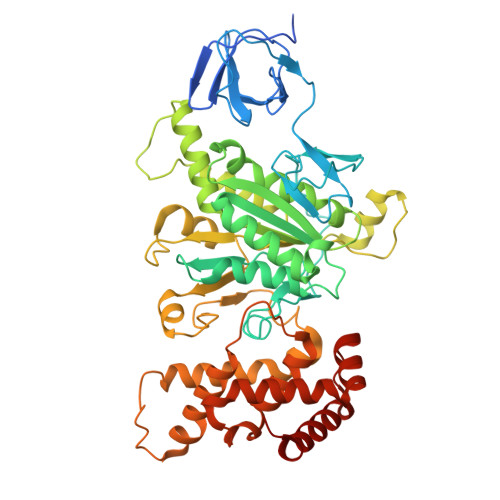
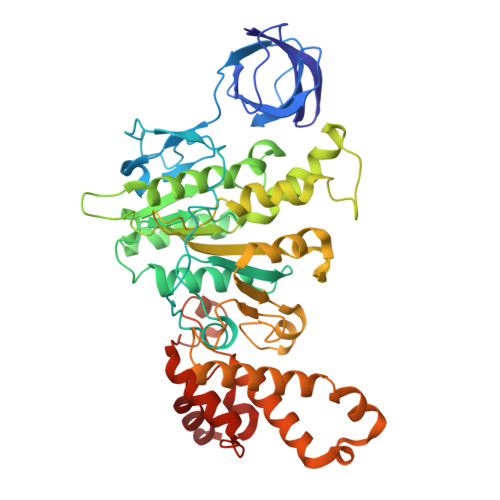
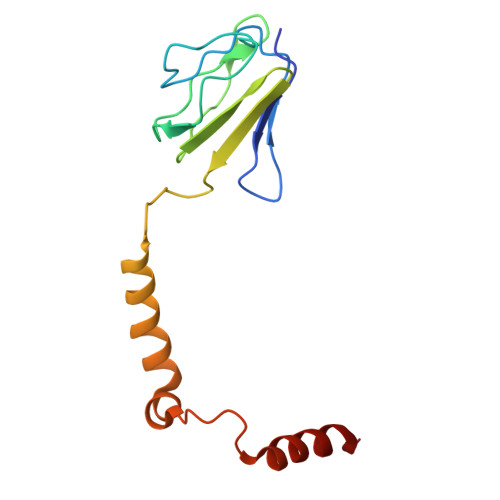
Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Le, K.C.M., Wong, C.F., Muller, V., Gruber, G.(2024) FASEB J 38: e70131-e70131
- PubMed: 39467208
- DOI: https://doi.org/10.1096/fj.202401629R
- Primary Citation of Related Structures:
8ZI0, 8ZI1, 8ZI2, 8ZI3 - PubMed Abstract:
Priority 1: critical WHO pathogen Acinetobacter baumannii depends on ATP synthesis and ATP:ADP homeostasis and its bifunctional F 1 F O -ATP synthase. While synthesizing ATP, it regulates ATP cleavage by its inhibitory ε subunit to prevent wasteful ATP consumption. We determined cryo-electron microscopy structures of the ATPase active A. baumannii F 1 -αßγε Δ134-139 mutant in four distinct conformational states, revealing four transition states and structural transformation of the ε's C-terminal domain, forming the switch of an ATP hydrolysis off- and an ATP synthesis on-state based. These alterations go in concert with altered motions and interactions in the catalytic- and rotary subunits of this engine. These A. baumannii interacting sites provide novel pathogen-specific targets for inhibitors, with the aim of ATP depletion and/or ATP synthesis and growth inhibition. Furthermore, the presented diversity to other bacterial F-ATP synthases extends the view of structural elements regulating such a catalyst.
- School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.
Organizational Affiliation: