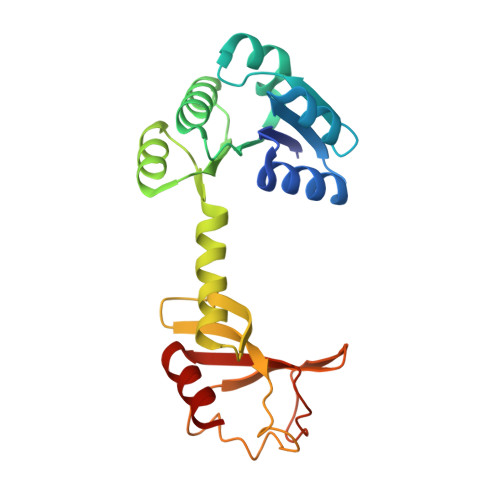
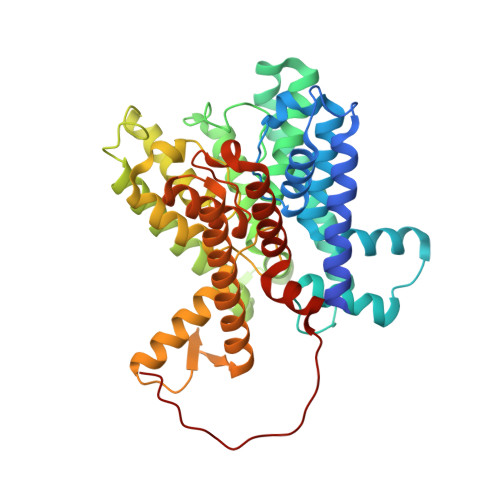
Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Chiang, W.T., Chang, Y.K., Hui, W.H., Chang, S.W., Liao, C.Y., Chang, Y.C., Chen, C.J., Wang, W.C., Lai, C.C., Wang, C.H., Luo, S.Y., Huang, Y.P., Chou, S.H., Horng, T.L., Hou, M.H., Muench, S.P., Chen, R.S., Tsai, M.D., Hu, N.J.(2024) Nat Commun 15: 3850-3850
- PubMed: 38719864
- DOI: https://doi.org/10.1038/s41467-024-48057-y
- Primary Citation of Related Structures:
8K16, 8K1K, 8K1S, 8K1T, 8K1U, 8XMH, 8XMI - PubMed Abstract:
The K + uptake system KtrAB is essential for bacterial survival in low K + environments. The activity of KtrAB is regulated by nucleotides and Na + . Previous studies proposed a putative gating mechanism of KtrB regulated by KtrA upon binding to ATP or ADP. However, how Na + activates KtrAB and the Na + binding site remain unknown. Here we present the cryo-EM structures of ATP- and ADP-bound KtrAB from Bacillus subtilis (BsKtrAB) both solved at 2.8 Å. A cryo-EM density at the intra-dimer interface of ATP-KtrA was identified as Na + , as supported by X-ray crystallography and ICP-MS. Thermostability assays and functional studies demonstrated that Na + binding stabilizes the ATP-bound BsKtrAB complex and enhances its K + flux activity. Comparing ATP- and ADP-BsKtrAB structures suggests that BsKtrB Arg417 and Phe91 serve as a channel gate. The synergism of ATP and Na + in activating BsKtrAB is likely applicable to Na + -activated K + channels in central nervous system.
- Graduate Institute of Biochemistry, National Chung Hsing University, Taichung, 402202, Taiwan.
Organizational Affiliation: