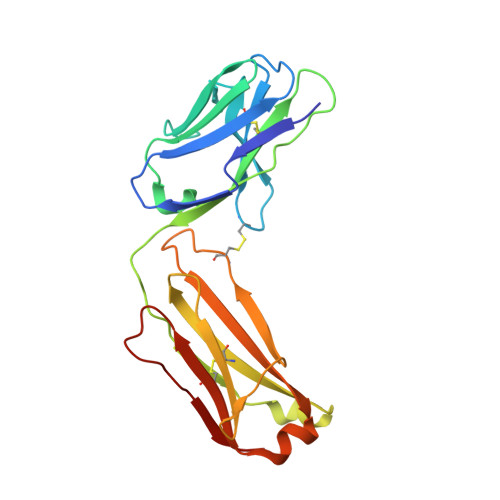
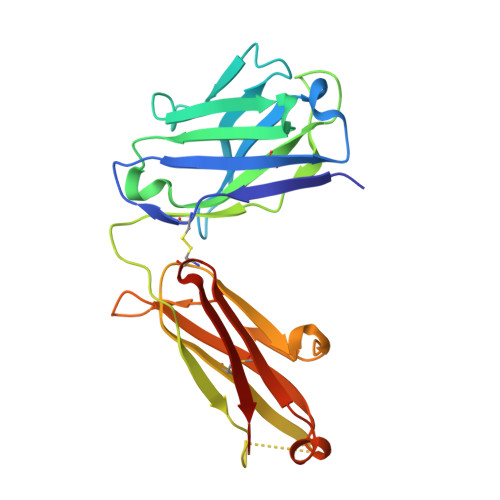
Stabilization of adalimumab Fab through the introduction of disulfide bonds between the variable and constant domains.
Yoshikawa, M., Senda, M., Nakamura, H., Oda-Ueda, N., Ueda, T., Senda, T., Ohkuri, T.(2024) Biochem Biophys Res Commun 700: 149592-149592
- PubMed: 38295648
- DOI: https://doi.org/10.1016/j.bbrc.2024.149592
- Primary Citation of Related Structures:
8XJ0 - PubMed Abstract:
Fab is a promising format for antibody drug. Therefore, efforts have been made to improve its thermal stability for therapeutic and commercial use. So far, we have attempted to introduce a disulfide bond into the Fab fragment to improve its thermal stability and demonstrated that it is possible to do this without sacrificing its biochemical function. In this study, to develop a novel stabilization strategy for Fab, we attempted to introduce a disulfide bond between the variable and constant domains and prepared three variants of Fab; H:G10C + H:P210C, L:P40C + L:E165C, and H:G10C + H:P210C + L:P40C + L:E165C. Differential scanning calorimetry measurements showed that each of these variants had improved thermal stability. In addition, the variants with two disulfide bonds demonstrated a 6.5 °C increase in their denaturation temperatures compared to wild-type Fab. The introduction of disulfide bonds was confirmed by X-ray crystallography, and the variants retained their antigen-binding activity. The variants were also found to be less aggregative than the wild type. Our results demonstrate that the introduction of a disulfide bond between the variable and constant domains significantly improves the thermal stability of Fab.
- Faculty of Pharmaceutical Sciences, Sojo University, Japan.
Organizational Affiliation: