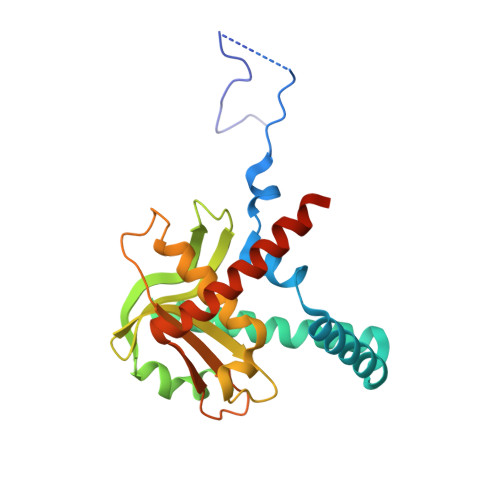
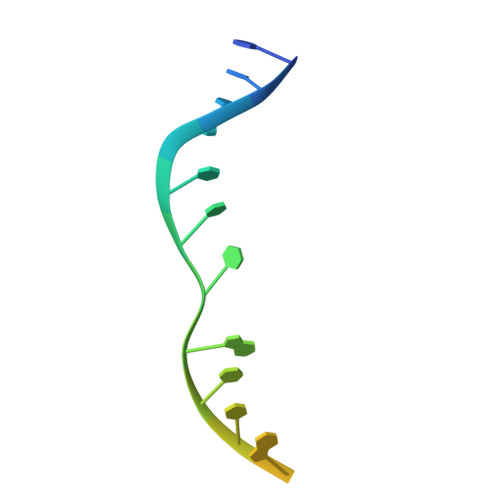
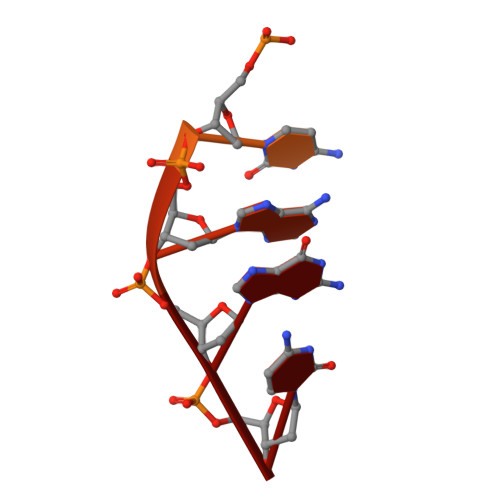
Structural basis of how MGME1 processes DNA 5' ends to maintain mitochondrial genome integrity.
Mao, E.Y.C., Yen, H.Y., Wu, C.C.(2024) Nucleic Acids Res 52: 4067-4078
- PubMed: 38471810
- DOI: https://doi.org/10.1093/nar/gkae186
- Primary Citation of Related Structures:
8XA9 - PubMed Abstract:
Mitochondrial genome maintenance exonuclease 1 (MGME1) helps to ensure mitochondrial DNA (mtDNA) integrity by serving as an ancillary 5'-exonuclease for DNA polymerase γ. Curiously, MGME1 exhibits unique bidirectionality in vitro, being capable of degrading DNA from either the 5' or 3' end. The structural basis of this bidirectionally and, particularly, how it processes DNA from the 5' end to assist in mtDNA maintenance remain unclear. Here, we present a crystal structure of human MGME1 in complex with a 5'-overhang DNA, revealing that MGME1 functions as a rigid DNA clamp equipped with a single-strand (ss)-selective arch, allowing it to slide on single-stranded DNA in either the 5'-to-3' or 3'-to-5' direction. Using a nuclease activity assay, we have dissected the structural basis of MGME1-derived DNA cleavage patterns in which the arch serves as a ruler to determine the cleavage site. We also reveal that MGME1 displays partial DNA-unwinding ability that helps it to better resolve 5'-DNA flaps, providing insights into MGME1-mediated 5'-end processing of nascent mtDNA. Our study builds on previously solved MGME1-DNA complex structures, finally providing the comprehensive functional mechanism of this bidirectional, ss-specific exonuclease.
- Department of Chemistry, College of Science, National Cheng Kung University, Tainan City 701, Taiwan.
Organizational Affiliation: