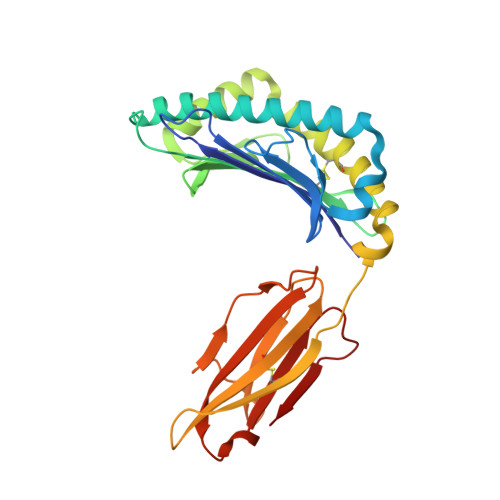
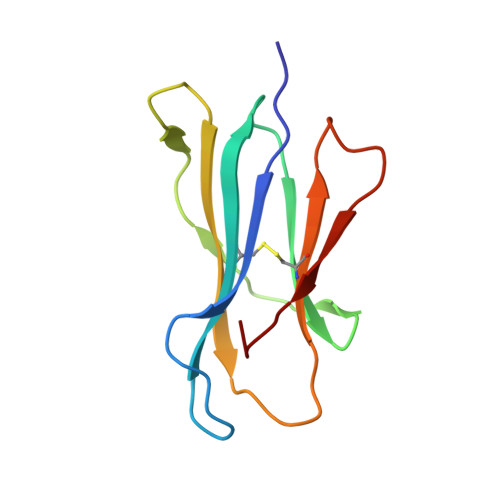
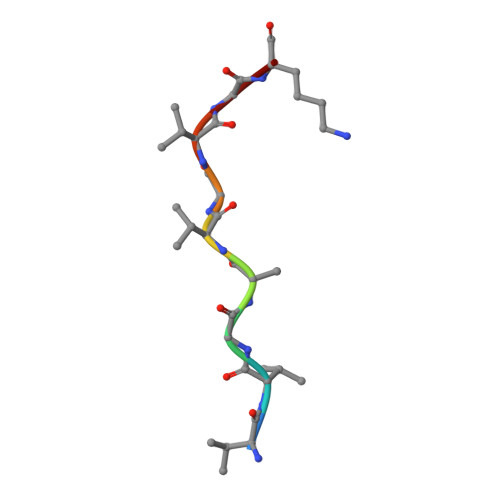
Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Zhang, M., Xu, W., Luo, L., Guan, F., Wang, X., Zhu, P., Zhang, J., Zhou, X., Wang, F., Ye, S.(2024) Commun Biol 7: 512-512
- PubMed: 38684865
- DOI: https://doi.org/10.1038/s42003-024-06209-2
- Primary Citation of Related Structures:
8WTE, 8WUL - PubMed Abstract:
Neoantigens derived from somatic mutations in Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS), the most frequently mutated oncogene, represent promising targets for cancer immunotherapy. Recent research highlights the potential role of human leukocyte antigen (HLA) allele A*11:01 in presenting these altered KRAS variants to the immune system. In this study, we successfully generate and identify murine T-cell receptors (TCRs) that specifically recognize KRAS 8-16 G12V from three predicted high affinity peptides. By determining the structure of the tumor-specific 4TCR2 bound to KRAS G12V -HLA-A*11:01, we conduct structure-based design to create and evaluate TCR variants with markedly enhanced affinity, up to 15.8-fold. This high-affinity TCR mutant, which involved only two amino acid substitutions, display minimal conformational alterations while maintaining a high degree of specificity for the KRAS G12V peptide. Our research unveils the molecular mechanisms governing TCR recognition towards KRAS G12V neoantigen and yields a range of affinity-enhanced TCR mutants with significant potential for immunotherapy strategies targeting tumors harboring the KRAS G12V mutation.
- Frontiers Science Center for Synthetic Biology (Ministry of Education), Tianjin Key Laboratory of Function and Application of Biological Macromolecular Structures, School of Life Sciences, Tianjin University, 92 Weijin Road, Nankai District, Tianjin, 300072, China.
Organizational Affiliation: