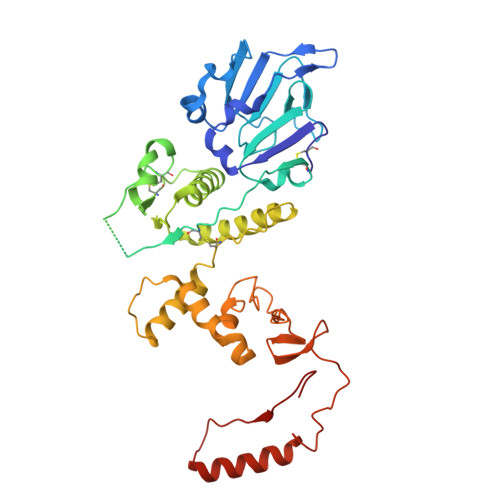
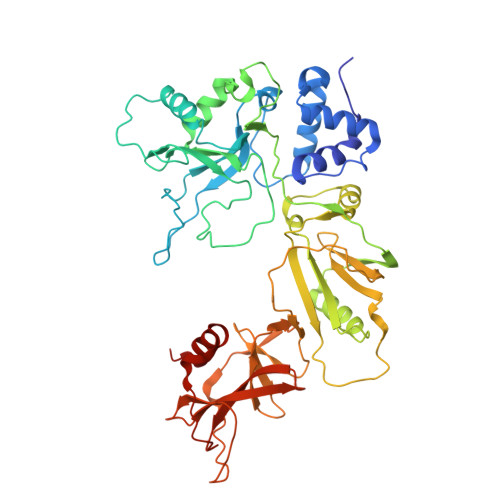
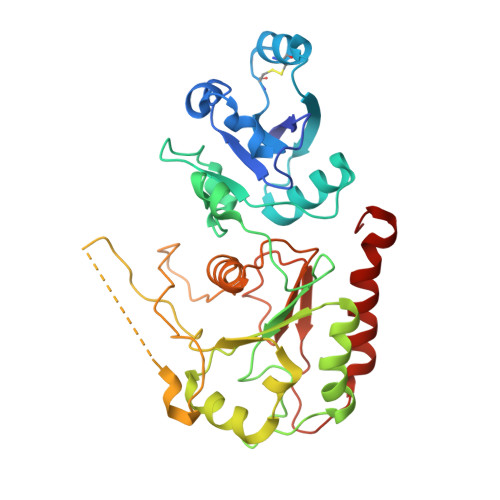
The nucleocapsid architecture and structural atlas of the prototype baculovirus define the hallmarks of a new viral realm.
Johnstone, B.A., Hardy, J.M., Ha, J., Butkovic, A., Koszalka, P., Accurso, C., Venugopal, H., de Marco, A., Krupovic, M., Coulibaly, F.(2024) Sci Adv 10: eado2631-eado2631
- PubMed: 39693434
- DOI: https://doi.org/10.1126/sciadv.ado2631
- Primary Citation of Related Structures:
8VWH, 8VWJ - PubMed Abstract:
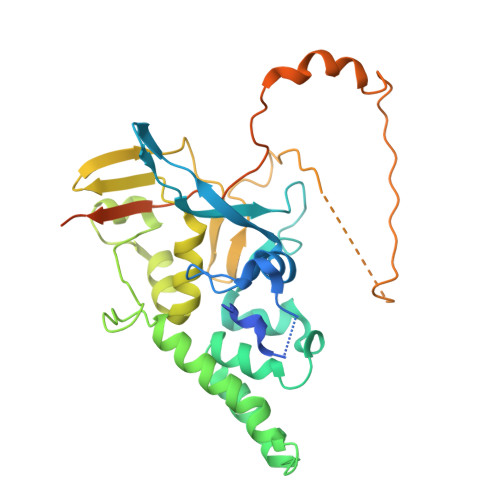
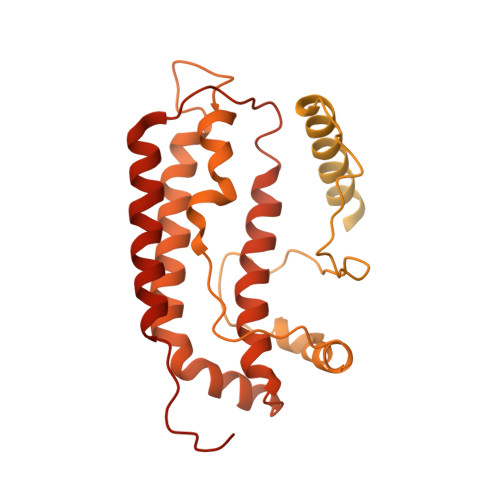
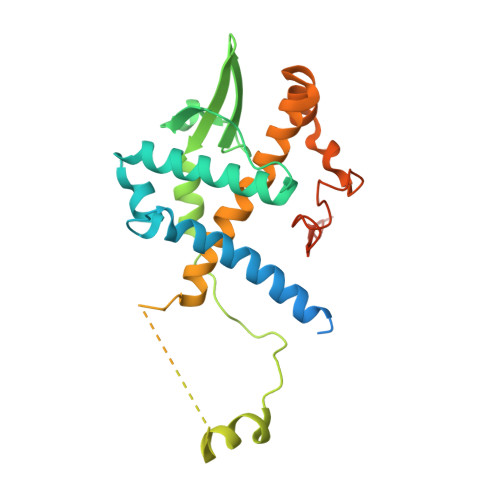
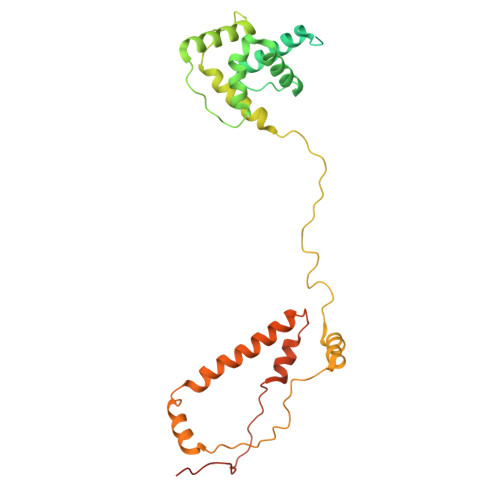
Baculovirus is the most studied insect virus owing to a broad ecological distribution and ease of engineering for biotechnological applications. However, its structure and evolutionary place in the virosphere remain enigmatic. Using cryo-electron microscopy, we show that the nucleocapsid forms a covalently cross-linked helical tube protecting a highly compacted 134-kilobase pair DNA genome. The ends of the tube are sealed by the base and cap substructures, which share a 126-subunit hub but differ in components that promote actin tail-mediated propulsion and nuclear entry of the nucleocapsid, respectively. Unexpectedly, sensitive searches for hidden evolutionary links show that the morphogenetic machinery and conserved oral infectivity factors originated within the lineage of baculo-like viruses (class Naldaviricetes ). The unique viral architecture and structural atlas of hallmark proteins firmly place these viruses into a separate new realm, the highest taxonomy rank, and provide a structural framework to expand their use as sustainable bioinsecticides and biomedical tools.
- Infection Program, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: