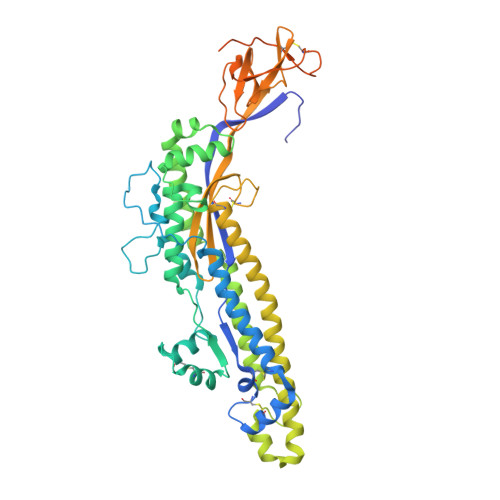
Simulation-driven design of stabilized SARS-CoV-2 spike S2 immunogens.
Nuqui, X., Casalino, L., Zhou, L., Shehata, M., Wang, A., Tse, A.L., Ojha, A.A., Kearns, F.L., Rosenfeld, M.A., Miller, E.H., Acreman, C.M., Ahn, S.H., Chandran, K., McLellan, J.S., Amaro, R.E.(2024) Nat Commun 15: 7370-7370
- PubMed: 39191724
- DOI: https://doi.org/10.1038/s41467-024-50976-9
- Primary Citation of Related Structures:
8VAO - PubMed Abstract:
The full-length prefusion-stabilized SARS-CoV-2 spike (S) is the principal antigen of COVID-19 vaccines. Vaccine efficacy has been impacted by emerging variants of concern that accumulate most of the sequence modifications in the immunodominant S1 subunit. S2, in contrast, is the most evolutionarily conserved region of the spike and can elicit broadly neutralizing and protective antibodies. Yet, S2's usage as an alternative vaccine strategy is hampered by its general instability. Here, we use a simulation-driven approach to design S2-only immunogens stabilized in a closed prefusion conformation. Molecular simulations provide a mechanistic characterization of the S2 trimer's opening, informing the design of tryptophan substitutions that impart kinetic and thermodynamic stabilization. Structural characterization via cryo-EM shows the molecular basis of S2 stabilization in the closed prefusion conformation. Informed by molecular simulations and corroborated by experiments, we report an engineered S2 immunogen that exhibits increased protein expression, superior thermostability, and preserved immunogenicity against sarbecoviruses.
- Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA, USA.
Organizational Affiliation: