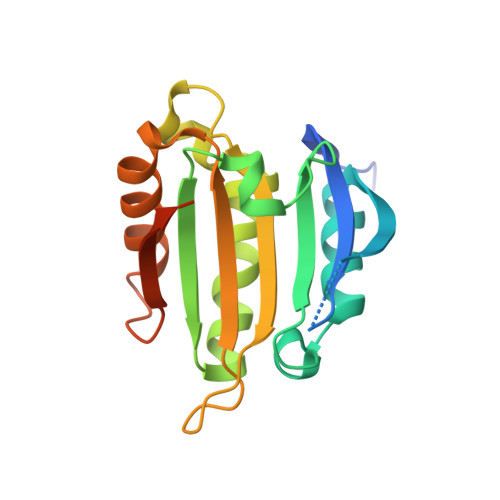
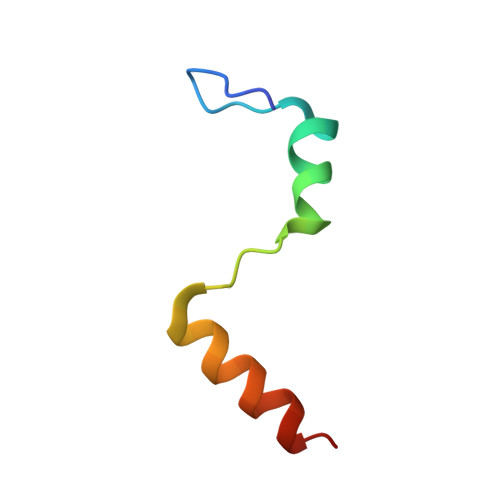
Structural analysis of the Trypanosoma brucei EIF4E6/EIF4G5 complex reveals details of the interaction between unusual eIF4F subunits.
Ferras Penteado, R., Santana da Silva, R., Nascimento Moura, D.M., Barbosa de Lima, G., Muniz Malvezzi, A., Tamara da Silva Monteiro, T., Cavalcanti Xavier, C., Vichier-Guerre, S., Dugue, L., Pochet, S., Tonin Zanchin, N.I., de Souza Reis, C.R., Pompilio de Melo Neto, O., Gomes Guimaraes, B.(2024) Sci Rep 14: 2178-2178
- PubMed: 38272944
- DOI: https://doi.org/10.1038/s41598-024-52364-1
- Primary Citation of Related Structures:
8UH1 - PubMed Abstract:
Recognition of the mRNA 5' end is a critical step needed for translation initiation. This step is performed by the cap binding protein eIF4E, which joins the larger eIF4G subunit to form the eIF4F complex. Trypanosomatids have a minimum of five different eIF4F-like complexes formed through specific but not well-defined interactions between four different eIF4E and five eIF4G homologues. The EIF4E6/EIF4G5 complex has been linked with the stage-specific translation of mRNAs encoding the major Trypanosoma brucei virulence factors. Here, to better define the molecular basis for the TbEIF4E6/TbEIF4G5 interaction, we describe the identification of the peptide interacting with TbEIF4E6 in the region comprising residues 79-166 of TbEIF4G5. The TbEIF4E6-TbEIF4G5_79-116 complex reconstituted with recombinant proteins is highly stable even in the absence of cap-4. The crystal structure of the complex was subsequently solved, revealing extensive interacting surfaces. Comparative analyses highlight the conservation of the overall structural arrangement of different eIF4E/eIF4G complexes. However, highly different interacting surfaces are formed with distinct binding contacts occurring both in the canonical and noncanonical elements within eIF4G and the respective eIF4E counterpart. These specific pairs of complementary interacting surfaces are likely responsible for the selective association needed for the formation of distinct eIF4F complexes in trypanosomatids.
- Carlos Chagas Institute - Oswaldo Cruz Foundation, Curitiba, PR, Brazil.
Organizational Affiliation: