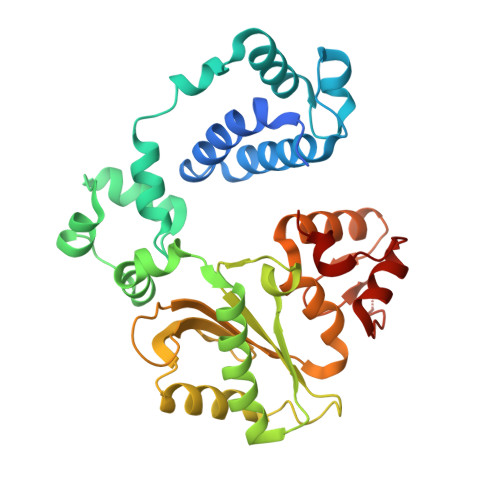
DNA polymerase lambda Loop1 variant yields unexpected gain-of-function capabilities in nonhomologous end-joining.
Kaminski, A.M., Chiruvella, K.K., Ramsden, D.A., Bebenek, K., Kunkel, T.A., Pedersen, L.C.(2024) DNA Repair (Amst) 136: 103645-103645
- PubMed: 38428373
- DOI: https://doi.org/10.1016/j.dnarep.2024.103645
- Primary Citation of Related Structures:
8U0O, 8U0P - PubMed Abstract:
DNA polymerases lambda (Polλ) and mu (Polμ) are X-Family polymerases that participate in DNA double-strand break (DSB) repair by the nonhomologous end-joining pathway (NHEJ). Both polymerases direct synthesis from one DSB end, using template derived from a second DSB end. In this way, they promote the NHEJ ligation step and minimize the sequence loss normally associated with this pathway. The two polymerases differ in cognate substrate, as Polλ is preferred when synthesis must be primed from a base-paired DSB end, while Polμ is required when synthesis must be primed from an unpaired DSB end. We generated a Polλ variant (Polλ KGET ) that retained canonical Polλ activity on a paired end-albeit with reduced incorporation fidelity. We recently discovered that the variant had unexpectedly acquired the activity previously unique to Polμ-synthesis from an unpaired primer terminus. Though the sidechains of the Loop1 region make no contact with the DNA substrate, Polλ KGET Loop1 amino acid sequence is surprisingly essential for its unique activity during NHEJ. Taken together, these results underscore that the Loop1 region plays distinct roles in different Family X polymerases.
- Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, 111 TW Alexander Dr., Bldg 101, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: