Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Meng, R., Xing, Z., Chang, J.Y., Yu, Z., Thongchol, J., Xiao, W., Wang, Y., Chamakura, K., Zeng, Z., Wang, F., Young, R., Zeng, L., Zhang, J.(2024) Nat Commun 15: 2746-2746
- PubMed: 38553443
- DOI: https://doi.org/10.1038/s41467-024-47119-5
- Primary Citation of Related Structures:
8TOB, 8TOC, 8TV9, 8TVA, 8TW2, 8TWC - PubMed Abstract:
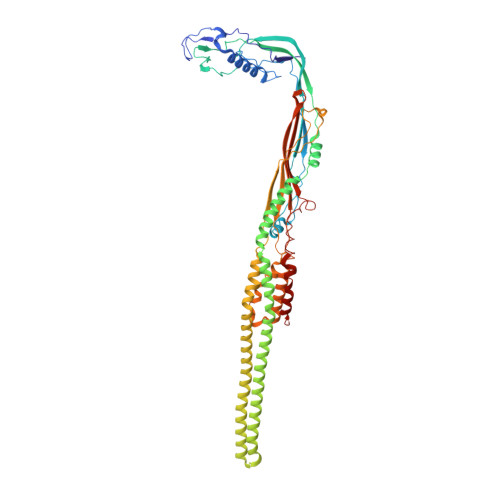
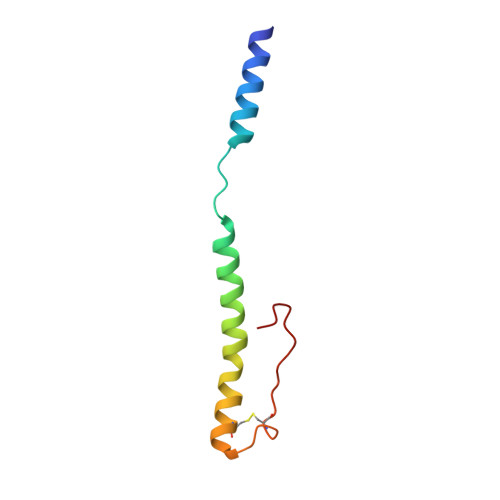
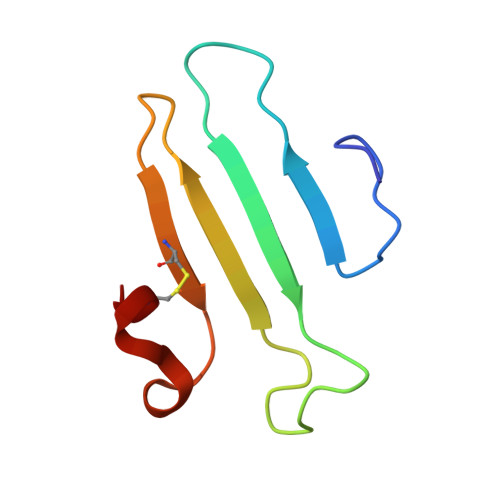
Acinetobacters pose a significant threat to human health, especially those with weakened immune systems. Type IV pili of acinetobacters play crucial roles in virulence and antibiotic resistance. Single-stranded RNA bacteriophages target the bacterial retractile pili, including type IV. Our study delves into the interaction between Acinetobacter phage AP205 and type IV pili. Using cryo-electron microscopy, we solve structures of the AP205 virion with an asymmetric dimer of maturation proteins, the native Acinetobacter type IV pili bearing a distinct post-translational pilin cleavage, and the pili-bound AP205 showing its maturation proteins adapted to pilin modifications, allowing each phage to bind to one or two pili. Leveraging these results, we develop a 20-kilodalton AP205-derived protein scaffold targeting type IV pili in situ, with potential for research and diagnostics.
- Center for Phage Technology, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX, 77843, USA.
Organizational Affiliation: