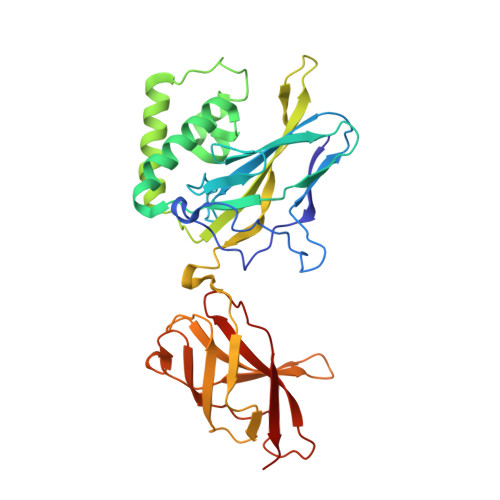
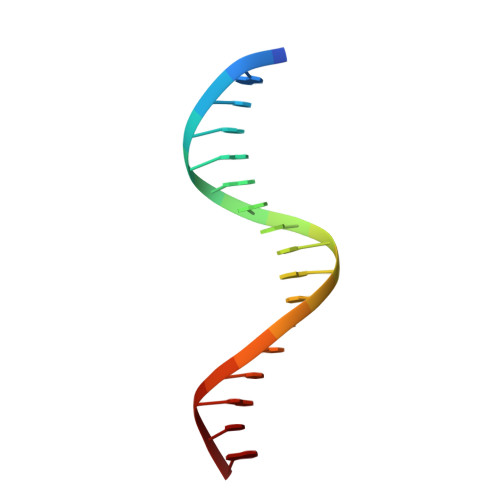
X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Zhu, N., Mealka, M., Mitchel, S., Milani, C., Acuna, L.M., Rogers, E., Lahana, A.N., Huxford, T.(2023) Biomolecules 13
- PubMed: 37759710
- DOI: https://doi.org/10.3390/biom13091310
- Primary Citation of Related Structures:
8TKL, 8TKM, 8TKN - PubMed Abstract:
Though originally characterized as an inactive or transcriptionally repressive factor, the NF-κB p50 homodimer has become appreciated as a physiologically relevant driver of specific target gene expression. By virtue of its low affinity for cytoplasmic IκB protein inhibitors, p50 accumulates in the nucleus of resting cells, where it is a binding target for the transcriptional co-activator IκBζ. In this study, we employed X-ray crystallography to analyze the structure of the p50 homodimer on κB DNA from the promoters of human interleukin-6 (IL-6) and neutrophil-gelatinase-associated lipocalin (NGAL) genes, both of which respond to IκBζ. The NF-κB p50 homodimer binds 11-bp on IL-6 κB DNA, while, on NGAL κB DNA, the spacing is 12-bp. This begs the question: what DNA binding mode is preferred by NF-κB p50 homodimer? To address this, we engineered a "Test" κB-like DNA containing the core sequence 5'-GGGGAATTCCCC-3' and determined its X-ray crystal structure in complex with p50. This revealed that, when presented with multiple options, NF-κB p50 homodimer prefers to bind 11-bp, which necessarily imposes asymmetry on the complex despite the symmetry inherent in both the protein and its target DNA, and that the p50 dimerization domain can contact DNA via distinct modes.
- Structural Biochemistry Laboratory, Department of Chemistry & Biochemistry, San Diego State University, 5500 Campanile Dr., San Diego, CA 92182-1030, USA.
Organizational Affiliation: