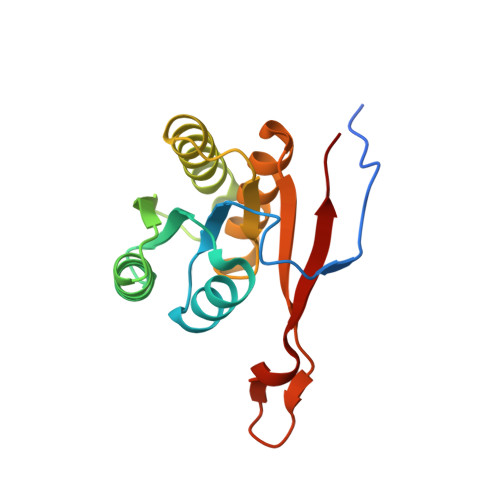
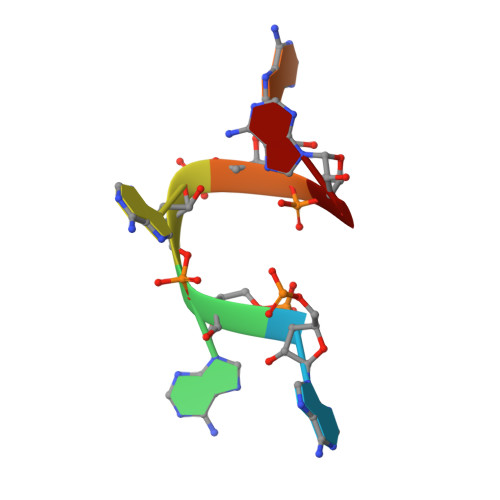
The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Baca, C.F., Yu, Y., Rostol, J.T., Majumder, P., Patel, D.J., Marraffini, L.A.(2024) Nature 625: 797-804
- PubMed: 38200316
- DOI: https://doi.org/10.1038/s41586-023-06902-y
- Primary Citation of Related Structures:
8T64, 8T65, 8T66 - PubMed Abstract:
Prokaryotic type III CRISPR-Cas systems provide immunity against viruses and plasmids using CRISPR-associated Rossman fold (CARF) protein effectors 1-5 . Recognition of transcripts of these invaders with sequences that are complementary to CRISPR RNA guides leads to the production of cyclic oligoadenylate second messengers, which bind CARF domains and trigger the activity of an effector domain 6,7 . Whereas most effectors degrade host and invader nucleic acids, some are predicted to contain transmembrane helices without an enzymatic function. Whether and how these CARF-transmembrane helix fusion proteins facilitate the type III CRISPR-Cas immune response remains unknown. Here we investigate the role of cyclic oligoadenylate-activated membrane protein 1 (Cam1) during type III CRISPR immunity. Structural and biochemical analyses reveal that the CARF domains of a Cam1 dimer bind cyclic tetra-adenylate second messengers. In vivo, Cam1 localizes to the membrane, is predicted to form a tetrameric transmembrane pore, and provides defence against viral infection through the induction of membrane depolarization and growth arrest. These results reveal that CRISPR immunity does not always operate through the degradation of nucleic acids, but is instead mediated via a wider range of cellular responses.
- Laboratory of Bacteriology, The Rockefeller University, New York, NY, USA.
Organizational Affiliation: