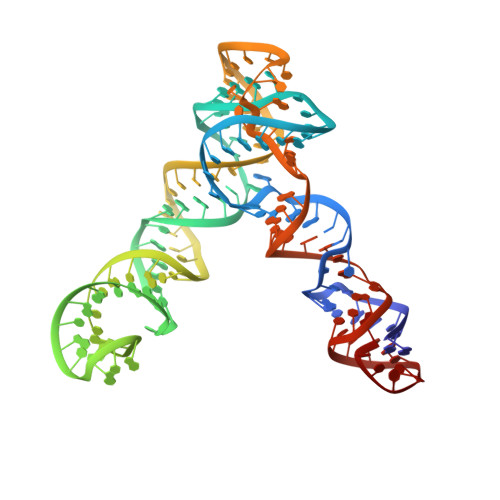
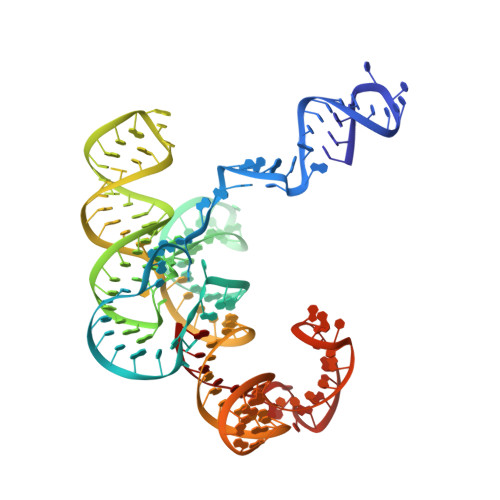
Cryo-EM structure and functional landscape of an RNA polymerase ribozyme.
McRae, E.K.S., Wan, C.J.K., Kristoffersen, E.L., Hansen, K., Gianni, E., Gallego, I., Curran, J.F., Attwater, J., Holliger, P., Andersen, E.S.(2024) Proc Natl Acad Sci U S A 121: e2313332121-e2313332121
- PubMed: 38207080
- DOI: https://doi.org/10.1073/pnas.2313332121
- Primary Citation of Related Structures:
8T2P - PubMed Abstract:
The emergence of an RNA replicase capable of self-replication is considered an important stage in the origin of life. RNA polymerase ribozymes (PR) - including a variant that uses trinucleotide triphosphates (triplets) as substrates - have been created by in vitro evolution and are the closest functional analogues of the replicase, but the structural basis for their function is poorly understood. Here we use single-particle cryogenic electron microscopy (cryo-EM) and high-throughput mutation analysis to obtain the structure of a triplet polymerase ribozyme (TPR) apoenzyme and map its functional landscape. The cryo-EM structure at 5-Å resolution reveals the TPR as an RNA heterodimer comprising a catalytic subunit and a noncatalytic, auxiliary subunit, resembling the shape of a left hand with thumb and fingers at a 70° angle. The two subunits are connected by two distinct kissing-loop (KL) interactions that are essential for polymerase function. Our combined structural and functional data suggest a model for templated RNA synthesis by the TPR holoenzyme, whereby heterodimer formation and KL interactions preorganize the TPR for optimal primer-template duplex binding, triplet substrate discrimination, and templated RNA synthesis. These results provide a better understanding of TPR structure and function and should aid the engineering of more efficient PRs.
- Interdisciplinary Nanoscience Center, Department of Molecular Biology and Genetics, Aarhus University, Aarhus 8000, Denmark.
Organizational Affiliation: