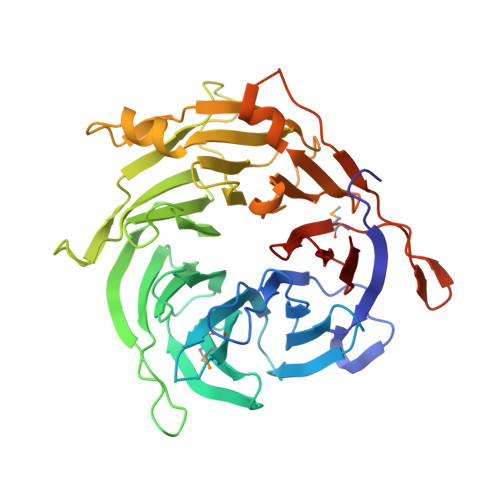
A dual role for the chromatin reader ORCA/LRWD1 in targeting the origin recognition complex to chromatin.
Sahu, S., Ekundayo, B.E., Kumar, A., Bleichert, F.(2023) EMBO J 42: e114654-e114654
- PubMed: 37551430
- DOI: https://doi.org/10.15252/embj.2023114654
- Primary Citation of Related Structures:
8SIU, 8SIY - PubMed Abstract:
Eukaryotic cells use chromatin marks to regulate the initiation of DNA replication. The origin recognition complex (ORC)-associated protein ORCA plays a critical role in heterochromatin replication in mammalian cells by recruiting the initiator ORC, but the underlying mechanisms remain unclear. Here, we report crystal and cryo-electron microscopy structures of ORCA in complex with ORC's Orc2 subunit and nucleosomes, establishing that ORCA orchestrates ternary complex assembly by simultaneously recognizing a highly conserved peptide sequence in Orc2, nucleosomal DNA, and repressive histone trimethylation marks through an aromatic cage. Unexpectedly, binding of ORCA to nucleosomes prevents chromatin array compaction in a manner that relies on H4K20 trimethylation, a histone modification critical for heterochromatin replication. We further show that ORCA is necessary and sufficient to specifically recruit ORC into chromatin condensates marked by H4K20 trimethylation, providing a paradigm for studying replication initiation in specific chromatin contexts. Collectively, our findings support a model in which ORCA not only serves as a platform for ORC recruitment to nucleosomes bearing specific histone marks but also helps establish a local chromatin environment conducive to subsequent MCM2-7 loading.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, USA.
Organizational Affiliation: