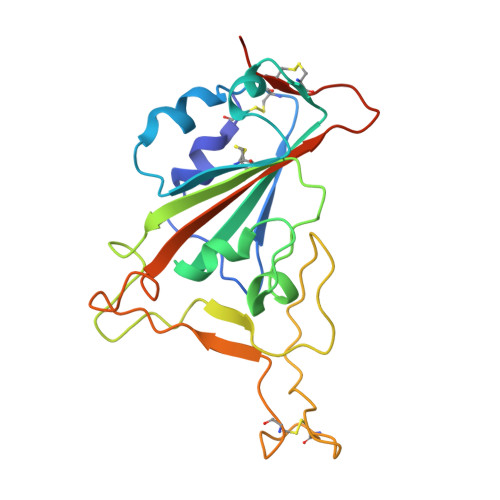
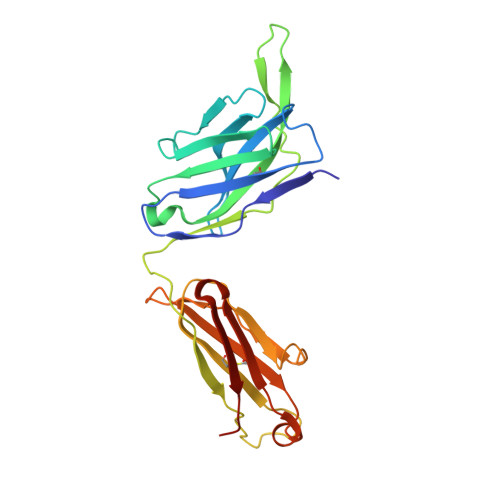
Broadly neutralizing antibodies targeting a conserved silent face of spike RBD resist extreme SARS-CoV-2 antigenic drift.
Song, G., Yuan, M., Liu, H., Capozzola, T., Lin, R.N., Torres, J.L., He, W.T., Musharrafieh, R., Dueker, K., Zhou, P., Callaghan, S., Mishra, N., Yong, P., Anzanello, F., Avillion, G., Vo, A.L., Li, X., Zhang, Y., Makhdoomi, M., Feng, Z., Zhu, X., Peng, L., Nemazee, D., Safonova, Y., Briney, B., Ward, A.B., Burton, D.R., Wilson, I.A., Andrabi, R.(2025) Cell Rep 44: 115948-115948
- PubMed: 40627497
- DOI: https://doi.org/10.1016/j.celrep.2025.115948
- Primary Citation of Related Structures:
8SIQ, 8SIR, 8SIS, 8SIT - PubMed Abstract:
Developing broad coronavirus vaccines hinges on identifying and understanding the molecular basis of conserved spike epitopes targeted by broadly neutralizing antibodies (bnAbs). Building on our earlier work identifying sarbecovirus receptor-binding domain (RBD) group 1 and 2 bnAbs, we now show that several of these antibodies retain neutralizing activity against highly mutated SARS-CoV-2 variants, including BA.2.86 and JN.1. Structural studies reveal that group 1 bnAbs use recurrent germline-encoded heavy-chain complementarity-determining region 3 (CDRH3) features to interact with a conserved RBD region that overlaps with class 4 bnAb site. Group 2 bnAbs recognize a less well-defined "site V" on the RBD and destabilize spike trimer. Notably, site V remains largely unchanged across SARS-CoV-2 variants and is conserved among diverse sarbecoviruses, highlighting its potential as a broad vaccine target. Our findings underscore the need for targeted vaccine strategies to induce immunofocused B cell responses to escape resistant subdominant spike RBD bnAb epitopes.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037, USA; IAVI Neutralizing Antibody Center, The Scripps Research Institute, La Jolla, CA 92037, USA; Consortium for HIV/AIDS Vaccine Development (CHAVD), The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: