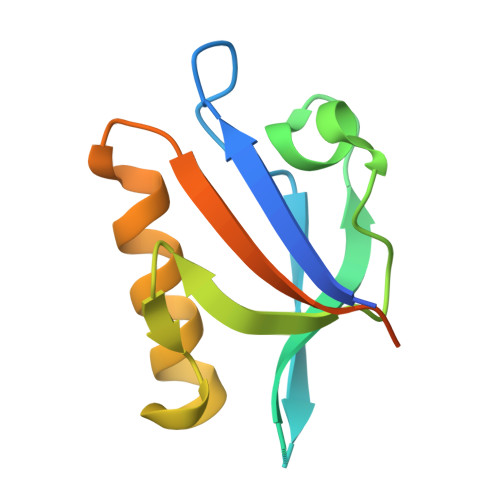
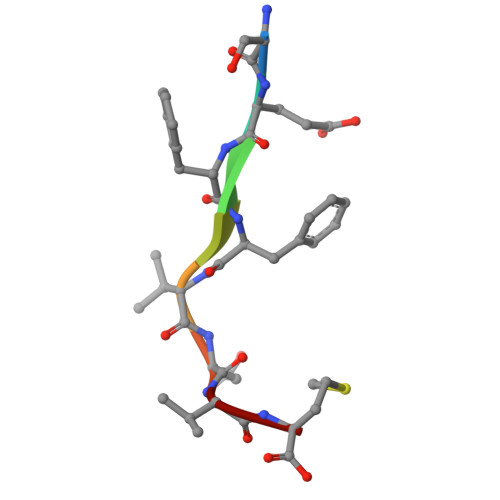
A class III ligand oscillates between internal and terminal binding modes as it engages with the Dishevelled PDZ domain.
Kumar, J., Micka, M., Komarek, J., Klumpler, T., Bystry, V., Sprangers, R., Barinka, C., Bryja, V., Tripsianes, K.(2025) Structure 33: 1362
- PubMed: 40516532
- DOI: https://doi.org/10.1016/j.str.2025.05.012
- Primary Citation of Related Structures:
8S6A - PubMed Abstract:
One of the largest domain-motif interactomes in human involves PSD-95/Discs-large/ZO-1 (PDZ) domains. The framework for understanding the PDZ interactome is well established; however the functional dynamics associated with PDZ-ligand interactions are poorly understood. Here, we report a dual PDZ-binding mode that ascribes unique dynamic features to class III ligand recognition. The crystal structure revealed that the PDZ domain can recognize either of the carboxylate moieties (terminal or internal) present in the class III ligand and laid out the register rules responsible for the dual recognition. Variants of the ligand designed to retain one or the other carboxylate of the native sequence were sufficient for PDZ binding. The conformational dynamics of PDZ probed by NMR relaxation dispersion experiments demonstrated that the class III ligand is shuffling binding modes as it engages with the PDZ domain. Our mechanistic findings reveal yet another aspect of PDZ binding plasticity specific to class III ligands.
- CEITEC - Central European Institute of Technology, Masaryk University, Brno, Czech Republic.
Organizational Affiliation: