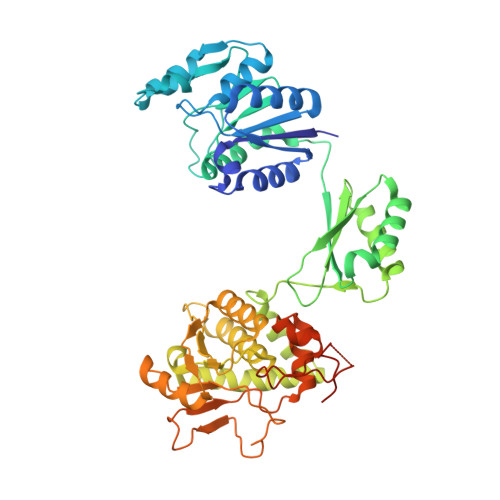
Structural insights into the bifunctional enzyme human FAD synthase.
Leo, G., Leone, P., Ataie Kachoie, E., Tolomeo, M., Galluccio, M., Indiveri, C., Barile, M., Capaldi, S.(2024) Structure 32: 953
- PubMed: 38688286
- DOI: https://doi.org/10.1016/j.str.2024.04.006
- Primary Citation of Related Structures:
8ROM, 8RON - PubMed Abstract:
Human flavin adenine dinucleotide synthase (hFADS) is a bifunctional, multi-domain enzyme that exhibits both flavin mononucleotide adenylyltransferase and pyrophosphatase activities. Here we report the crystal structure of full-length hFADS2 and its C-terminal PAPS domain in complex with flavin adenine dinucleotide (FAD), and dissect the structural determinants underlying the contribution of each individual domain, within isoforms 1 and 2, to each of the two enzymatic activities. Structural and functional characterization performed on complete or truncated constructs confirmed that the C-terminal domain tightly binds FAD and catalyzes its synthesis, while the combination of the N-terminal molybdopterin-binding and KH domains is the minimal essential substructure required for the hydrolysis of FAD and other ADP-containing dinucleotides. hFADS2 associates in a stable C2-symmetric dimer, in which the packing of the KH domain of one protomer against the N-terminal domain of the other creates the adenosine-specific active site responsible for the hydrolytic activity.
- Department of Biotechnology, University of Verona, Strada Le Grazie 15, 37134 Verona, Italy.
Organizational Affiliation: