Antibody-mediated TGF-beta 1 activation for the treatment of diseases caused by deleterious T cell activity.
Lambert, F., Felix, J., Wautier, S., Dupre, E., Jamez, M., Michiels, C., Gaignage, M., Marien, L., Lesage, M., van der Woning, B., Savvides, S.N., Lucas, S.(2025) Cell Rep 44: 116061-116061
- PubMed: 40742810
- DOI: https://doi.org/10.1016/j.celrep.2025.116061
- Primary Citation of Related Structures:
8REW, 8REX - PubMed Abstract:
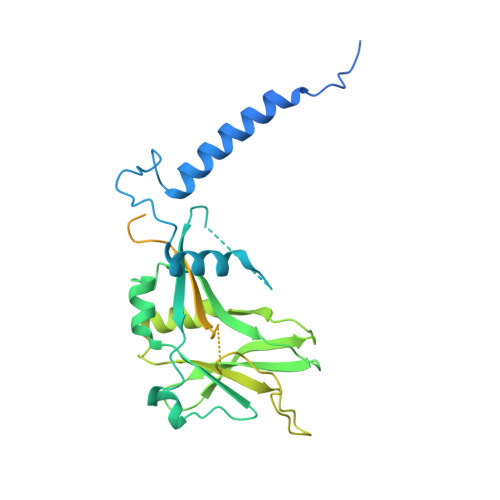
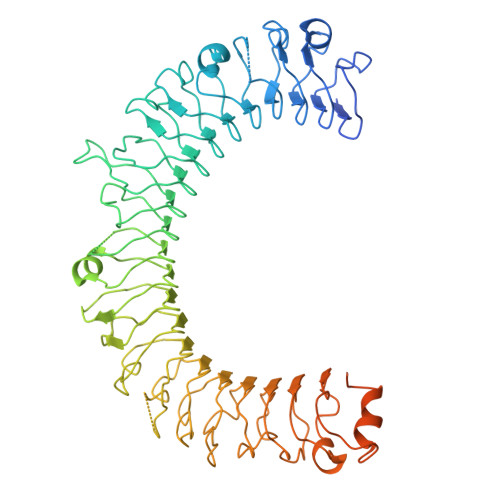
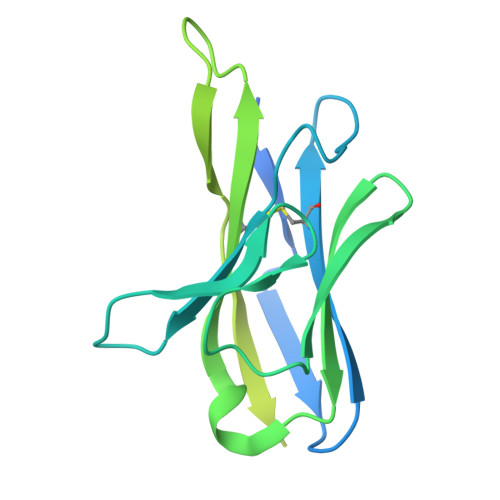
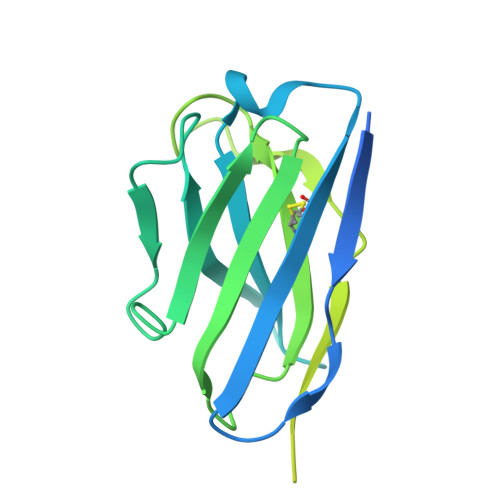
Transforming growth factor β1 (TGF-β1) is an immunosuppressive cytokine produced as a latent homodimer, in which mature TGF-β1 is encapsulated and kept inactive by the latency-associated peptide (LAP). The transmembrane protein GARP presents latent TGF-β1 on the surface of regulatory T cells (Tregs) to enable activation and release of mature TGF-β1 by integrins. Here, we derived monoclonal antibodies (mAbs) that activate latent TGF-β1 anchored on cells by a transmembrane protein. Biochemical and structural studies by electron cryo-microscopy (cryo-EM) reveal that such mAb-mediated activation requires bivalent binding close to the LAP dimerization interface and crosslinking of two membrane-bound GARP:TGF-β1 complexes on the same cell or across different cells. Administration of mAbs to mice with graft versus host disease reduced disease severity and increased survival. The therapeutic effect required Tregs. Collectively, our findings demonstrate that activation of membrane-bound TGF-β1 in vivo is achievable with mAbs, introducing new immunotherapeutic options for allo- or autoimmune diseases characterized by deleterious T cell activity insufficiently controlled by Tregs.
- de Duve Institute, UCLouvain, 1200 Brussels, Belgium.
Organizational Affiliation: