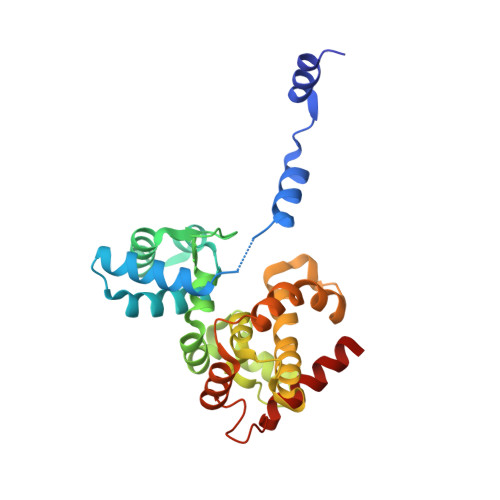
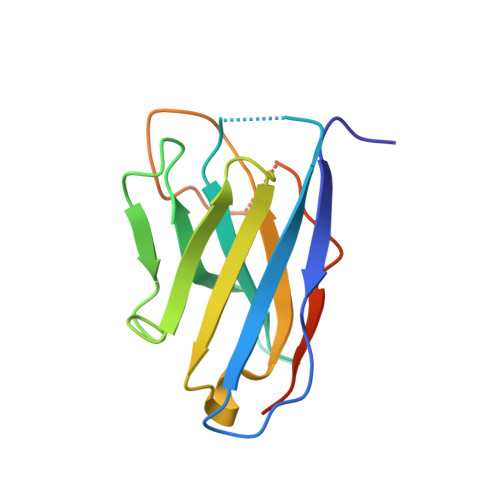
Structural flexibility of Toscana virus nucleoprotein in the presence of a single-chain camelid antibody.
Papageorgiou, N., Baklouti, A., Lichiere, J., Desmyter, A., Canard, B., Coutard, B., Ferron, F.(2024) Acta Crystallogr D Struct Biol 80: 113-122
- PubMed: 38265877
- DOI: https://doi.org/10.1107/S2059798324000196
- Primary Citation of Related Structures:
8RCQ - PubMed Abstract:
Phenuiviridae nucleoprotein is the main structural and functional component of the viral cycle, protecting the viral RNA and mediating the essential replication/transcription processes. The nucleoprotein (N) binds the RNA using its globular core and polymerizes through the N-terminus, which is presented as a highly flexible arm, as demonstrated in this article. The nucleoprotein exists in an `open' or a `closed' conformation. In the case of the closed conformation the flexible N-terminal arm folds over the RNA-binding cleft, preventing RNA adsorption. In the open conformation the arm is extended in such a way that both RNA adsorption and N polymerization are possible. In this article, single-crystal X-ray diffraction and small-angle X-ray scattering were used to study the N protein of Toscana virus complexed with a single-chain camelid antibody (VHH) and it is shown that in the presence of the antibody the nucleoprotein is unable to achieve a functional assembly to form a ribonucleoprotein complex.
- Université Aix-Marseille, Architecture et Fonction des Macromolécules Biologiques (AFMB)-UMR7257 CNRS, Case 925, 163 Avenue de Luminy, 13009 Marseille, France.
Organizational Affiliation: