Substrate recruitment via eIF2 gamma enhances catalytic efficiency of a holophosphatase that terminates the integrated stress response.
Yan, Y., Shetty, M., Harding, H.P., George, G., Zyryanova, A., Labbe, K., Mafi, A., Hao, Q., Sidrauski, C., Ron, D.(2024) Proc Natl Acad Sci U S A 121: e2320013121-e2320013121
- PubMed: 38547060
- DOI: https://doi.org/10.1073/pnas.2320013121
- Primary Citation of Related Structures:
8QZZ - PubMed Abstract:
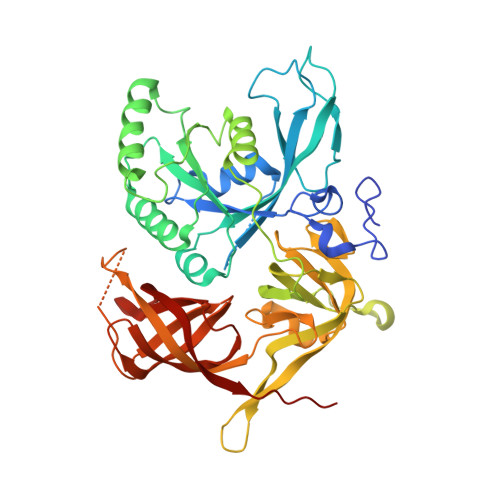
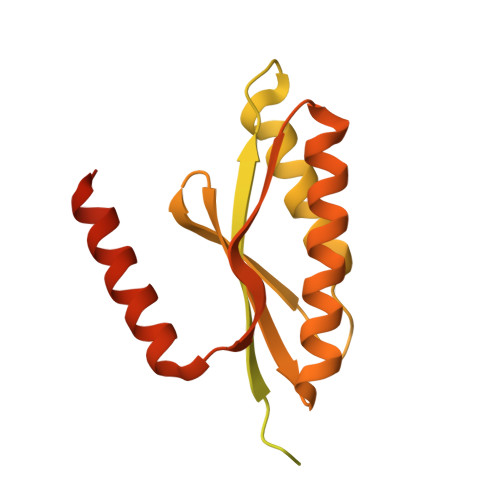
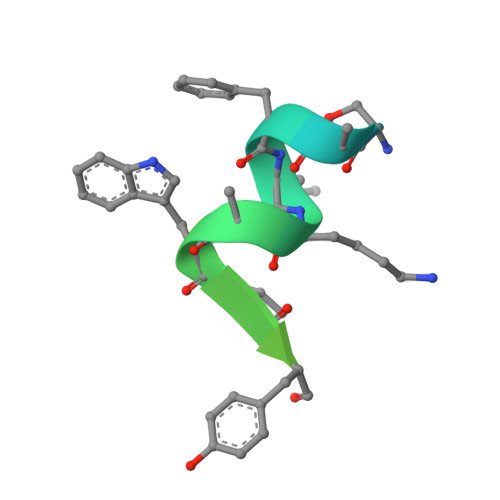
Dephosphorylation of pSer51 of the α subunit of translation initiation factor 2 (eIF2α P ) terminates signaling in the integrated stress response (ISR). A trimeric mammalian holophosphatase comprised of a protein phosphatase 1 (PP1) catalytic subunit, the conserved C-terminally located ~70 amino acid core of a substrate-specific regulatory subunit (PPP1R15A/GADD34 or PPP1R15B/CReP) and G-actin (an essential cofactor) efficiently dephosphorylate eIF2α P in vitro. Unlike their viral or invertebrate counterparts, with whom they share the conserved 70 residue core, the mammalian PPP1R15s are large proteins of more than 600 residues. Genetic and cellular observations point to a functional role for regions outside the conserved core of mammalian PPP1R15A in dephosphorylating its natural substrate, the eIF2 trimer. We have combined deep learning technology, all-atom molecular dynamics simulations, X-ray crystallography, and biochemistry to uncover binding of the γ subunit of eIF2 to a short helical peptide repeated four times in the functionally important N terminus of human PPP1R15A that extends past its conserved core. Binding entails insertion of Phe and Trp residues that project from one face of an α-helix formed by the conserved repeats of PPP1R15A into a hydrophobic groove exposed on the surface of eIF2γ in the eIF2 trimer. Replacing these conserved Phe and Trp residues with Ala compromises PPP1R15A function in cells and in vitro. These findings suggest mechanisms by which contacts between a distant subunit of eIF2 and elements of PPP1R15A distant to the holophosphatase active site contribute to dephosphorylation of eIF2α P by the core PPP1R15 holophosphatase and to efficient termination of the ISR in mammals.
- Cambridge Institute for Medical Research, Department of Clinical Biochemistry, University of Cambridge, Cambridge CB2 0XY, United Kingdom.
Organizational Affiliation: