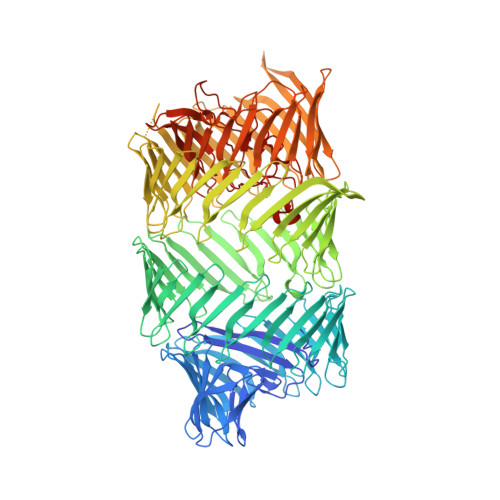
T6SS-associated Rhs toxin-encapsulating shells: Structural and bioinformatical insights into bacterial weaponry and self-protection.
Kielkopf, C.S., Shneider, M.M., Leiman, P.G., Taylor, N.M.I.(2024) Structure 32: 2375
- PubMed: 39481373
- DOI: https://doi.org/10.1016/j.str.2024.10.008
- Primary Citation of Related Structures:
8QJA, 8QJB, 8QJC, 8QJD - PubMed Abstract:
Bacteria use the type VI secretion system (T6SS) to secrete toxins into pro- and eukaryotic cells via machinery consisting of a contractile sheath and a rigid tube. Rearrangement hotspot (Rhs) proteins represent one of the most common T6SS effectors. The Rhs C-terminal toxin domain displays great functional diversity, while the Rhs core is characterized by YD repeats. We elucidate the Rhs core structures of PAAR- and VgrG-linked Rhs proteins from Salmonella bongori and Advenella mimigardefordensis, respectively. The Rhs core forms a large shell of β-sheets with a negatively charged interior and encloses a large volume. The S. bongori Rhs toxin does not lead to ordered density in the Rhs shell, suggesting the toxin is unfolded. Together with bioinformatics analysis showing that Rhs toxins predominantly act intracellularly, this suggests that the Rhs core functions two-fold, as a safety feature for the producer cell and as delivery mechanism for the toxin.
- Structural Biology of Molecular Machines Group, Protein Structure & Function Program, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200 Copenhagen, Denmark.
Organizational Affiliation: