Genome anchoring, retention, and release by neck proteins of Staphylococcus phage 812.
Cienikova, Z., Novacek, J., Siborova, M., Popelarova, B., Fuzik, T., Botka, T., Benesik, M., Bardy, P., Pantucek, R., Plevka, P.(2026) Commun Biol
- PubMed: 41507424
- DOI: https://doi.org/10.1038/s42003-025-09477-8
- Primary Citation of Related Structures:
8Q01, 8Q1I, 8Q7D, 8QEK, 8QEM, 8QGR, 8QJE, 8QKH, 8R5G, 8R69 - PubMed Abstract:
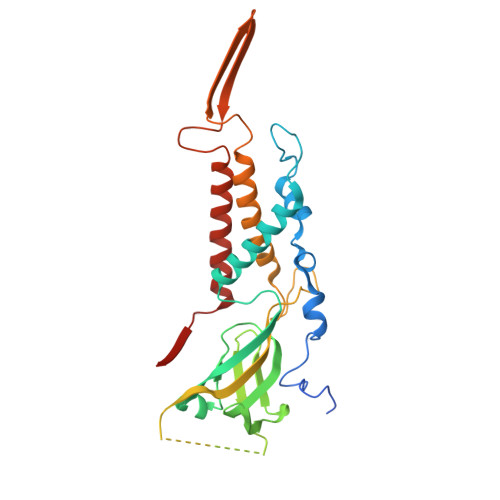
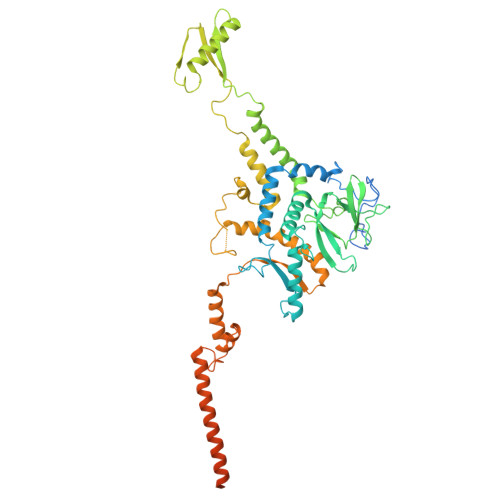
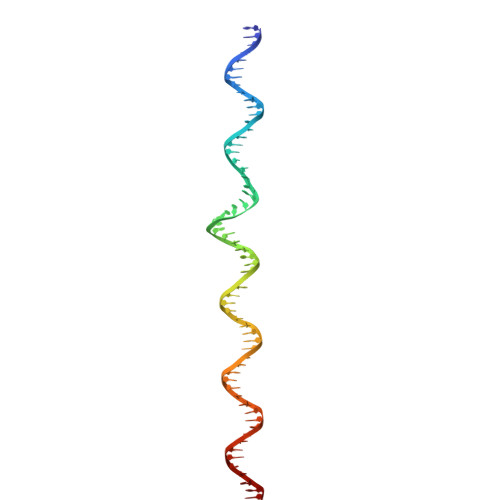
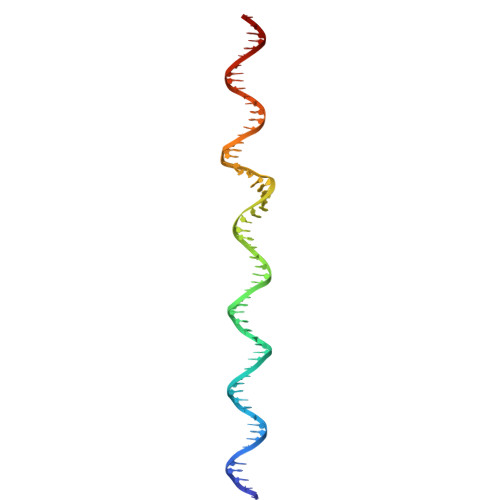
The virion of Staphylococcus phage 812 is formed by a capsid and a contractile tail joined together by neck proteins. The neck proteins are crucial for virion assembly, DNA packaging, and the regulation of genome release, but their functions are not completely understood. Here, we show that the neck of phage 812 consists of portal, adaptor, stopper, tail terminator, and two types of decoration proteins. A dodecameric DNA-binding site at the surface of the portal complex anchors the phage genome inside the capsid. The adaptor complex induces a local B-to-A form transition of the DNA in the neck channel that could slow or pause genome translocation during ejection. The central channel of a stopper complex that is not attached to the tail terminator complex is closed by gating loops. In contrast, in the phage 812 virion, the gating loops are in an open conformation, and the DNA extends into the tail. The structure of neck proteins is not affected by tail sheath contraction. Therefore, the expulsion of tail tape measure proteins triggers the genome release.
- CEITEC Masaryk University, Kamenice 753/5, 625 00, Brno, Czech Republic.
Organizational Affiliation: