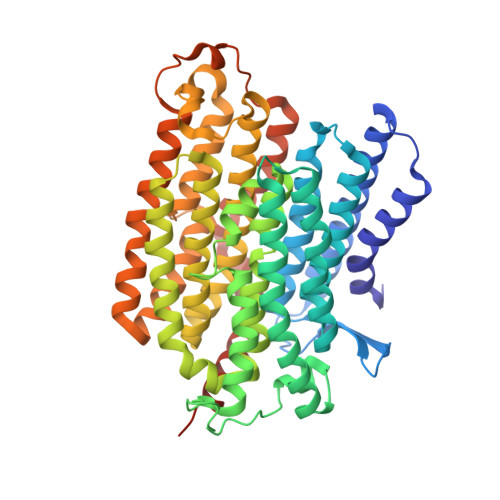
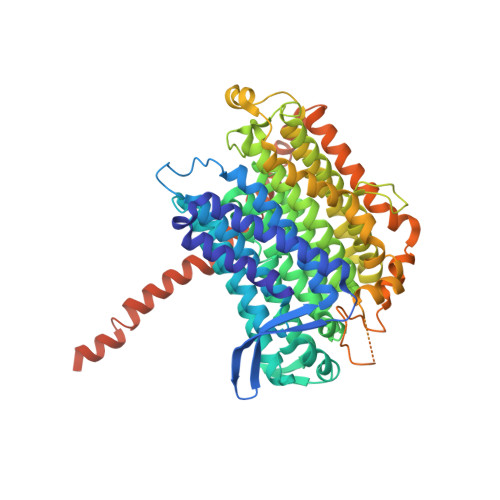
Structure of the turnover-ready state of an ancestral respiratory complex I.
Ivanov, B.S., Bridges, H.R., Jarman, O.D., Hirst, J.(2024) Nat Commun 15: 9340-9340
- PubMed: 39472559
- DOI: https://doi.org/10.1038/s41467-024-53679-3
- Primary Citation of Related Structures:
8QBY, 8QC1 - PubMed Abstract:
Respiratory complex I is pivotal for cellular energy conversion, harnessing energy from NADH:ubiquinone oxidoreduction to drive protons across energy-transducing membranes for ATP synthesis. Despite detailed structural information on complex I, its mechanism of catalysis remains elusive due to lack of accompanying functional data for comprehensive structure-function analyses. Here, we present the 2.3-Å resolution structure of complex I from the α-proteobacterium Paracoccus denitrificans, a close relative of the mitochondrial progenitor, in phospholipid-bilayer nanodiscs. Three eukaryotic-type supernumerary subunits (NDUFS4, NDUFS6 and NDUFA12) plus a novel L-isoaspartyl-O-methyltransferase are bound to the core complex. Importantly, the enzyme is in a single, homogeneous resting state that matches the closed, turnover-ready (active) state of mammalian complex I. Our structure reveals the elements that stabilise the closed state and completes P. denitrificans complex I as a unified platform for combining structure, function and genetics in mechanistic studies.
- The Medical Research Council Mitochondrial Biology Unit, University of Cambridge, Keith Peters Building, Cambridge Biomedical Campus, Cambridge, UK.
Organizational Affiliation: