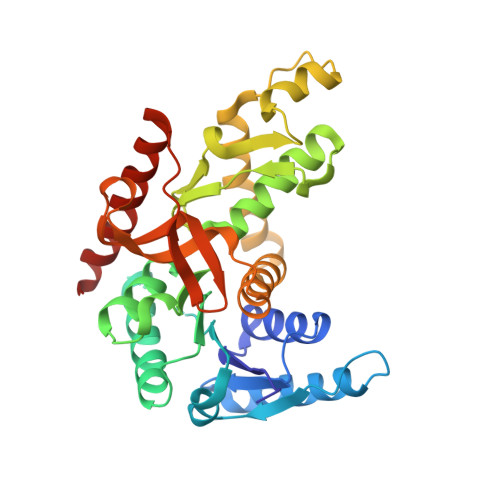
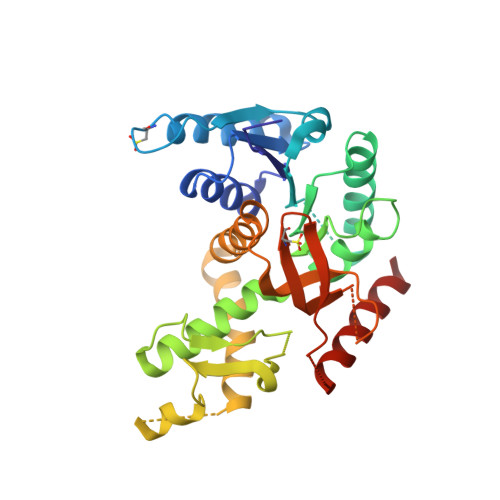
Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
Bertrand, Q., Coquille, S., Iorio, A., Sterpone, F., Madern, D.(2023) J Struct Biol 215: 108039-108039
- PubMed: 37884067
- DOI: https://doi.org/10.1016/j.jsb.2023.108039
- Primary Citation of Related Structures:
7NAY, 8Q3C - PubMed Abstract:
In this work, we investigated the lactate dehydrogenase (LDH) from Selenomonas ruminantium (S. rum), an enzyme that differs at key amino acid positions from canonical allosteric LDHs. The wild type (Wt) of this enzyme recognises pyuvate as all LDHs. However, introducing a single point mutation in the active site loop (I85R) allows S. Rum LDH to recognize the oxaloacetate substrate as a typical malate dehydrogenase (MalDH), whilst maintaining homotropic activation as an LDH. We report the tertiary structure of the Wt and I85RLDH mutant. The Wt S. rum enzyme structure binds NADH and malonate, whilst also resembling the typical compact R-active state of canonical LDHs. The structure of the mutant with I85R was solved in the Apo State (without ligand), and shows no large conformational reorganization such as that observed with canonical allosteric LDHs in Apo state. This is due to a local structural feature typical of S. rum LDH that prevents large-scale conformational reorganization. The S. rum LDH was also studied using Molecular Dynamics simulations, probing specific local deformations of the active site that allow the S. rum LDH to sample the T-inactive state. We propose that, with respect to the LDH/MalDH superfamily, the S. rum enzyme possesses a specificstructural and dynamical way to ensure homotropic activation.
- Univ. Grenoble Alpes, CEA, CNRS, IBS, 38000 Grenoble, France; Laboratory of Biomolecular Research, Biology and Chemistry Division, Paul Scherrer Institut, Villigen, Switzerland.
Organizational Affiliation: